Mapping hydration dynamics around a protein surface
- PMID: 18003912
- PMCID: PMC2141799
- DOI: 10.1073/pnas.0707647104
Mapping hydration dynamics around a protein surface
Abstract
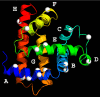
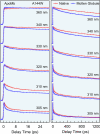
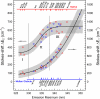
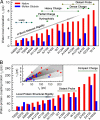
Protein surface hydration is fundamental to its structure and activity. We report here the direct mapping of global hydration dynamics around a protein in its native and molten globular states, using a tryptophan scan by site-specific mutations. With 16 tryptophan mutants and in 29 different positions and states, we observed two robust, distinct water dynamics in the hydration layer on a few ( approximately 1-8 ps) and tens to hundreds of picoseconds ( approximately 20-200 ps), representing the initial local relaxation and subsequent collective network restructuring, respectively. Both time scales are strongly correlated with protein's structural and chemical properties. These results reveal the intimate relationship between hydration dynamics and protein fluctuations and such biologically relevant water-protein interactions fluctuate on picosecond time scales.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Pal SK, Zewail AH. Chem Rev. 2004;104:2099–2123. - PubMed
-
- Wüthrich K. Angew Chem Int Ed. 2003;42:3340–3363. - PubMed
-
- Levy Y, Onuchic JN. Annu Rev Biophys Biomol Struct. 2006;35:389–415. - PubMed
-
- Mattos C. Trends Biochem Sci. 2002;27:203–208. - PubMed
-
- Garczarek F, Gerwert K. Nature. 2006;439:109–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

