Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer
- PMID: 18003927
- PMCID: PMC2141832
- DOI: 10.1073/pnas.0704652104
Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer
Abstract
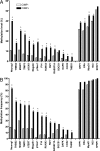
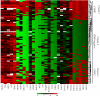
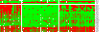
Colon cancer has been viewed as the result of progressive accumulation of genetic and epigenetic abnormalities. However, this view does not fully reflect the molecular heterogeneity of the disease. We have analyzed both genetic (mutations of BRAF, KRAS, and p53 and microsatellite instability) and epigenetic alterations (DNA methylation of 27 CpG island promoter regions) in 97 primary colorectal cancer patients. Two clustering analyses on the basis of either epigenetic profiling or a combination of genetic and epigenetic profiling were performed to identify subclasses with distinct molecular signatures. Unsupervised hierarchical clustering of the DNA methylation data identified three distinct groups of colon cancers named CpG island methylator phenotype (CIMP) 1, CIMP2, and CIMP negative. Genetically, these three groups correspond to very distinct profiles. CIMP1 are characterized by MSI (80%) and BRAF mutations (53%) and rare KRAS and p53 mutations (16% and 11%, respectively). CIMP2 is associated with 92% KRAS mutations and rare MSI, BRAF, or p53 mutations (0, 4, and 31% respectively). CIMP-negative cases have a high rate of p53 mutations (71%) and lower rates of MSI (12%) or mutations of BRAF (2%) or KRAS (33%). Clustering based on both genetic and epigenetic parameters also identifies three distinct (and homogeneous) groups that largely overlap with the previous classification. The three groups are independent of age, gender, or stage, but CIMP1 and 2 are more common in proximal tumors. Together, our integrated genetic and epigenetic analysis reveals that colon cancers correspond to three molecularly distinct subclasses of disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
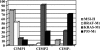

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

