Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane
- PMID: 18003975
- PMCID: PMC2174176
- DOI: 10.1091/mbc.e07-08-0748
Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane
Abstract
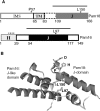
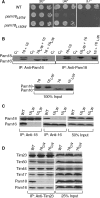
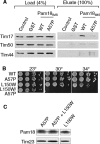
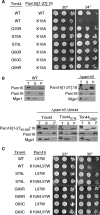
Import of proteins across the inner mitochondrial membrane through the Tim23:Tim17 translocase requires the function of an essential import motor having mitochondrial 70-kDa heat-shock protein (mtHsp70) at its core. The heterodimer composed of Pam18, the J-protein partner of mtHsp70, and the related protein Pam16 is a critical component of this motor. We report that three interactions contribute to association of the heterodimer with the translocon: the N terminus of Pam16 with the matrix side of the translocon, the inner membrane space domain of Pam18 (Pam18(IMS)) with Tim17, and the direct interaction of the J-domain of Pam18 with the J-like domain of Pam16. Pam16 plays a major role in translocon association, as alterations affecting the stability of the Pam18:Pam16 heterodimer dramatically affect association of Pam18, but not Pam16, with the translocon. Suppressors of the growth defects caused by alterations in the N terminus of Pam16 were isolated and found to be due to mutations in a short segment of TIM44, the gene encoding the peripheral membrane protein that tethers mtHsp70 to the translocon. These data suggest a model in which Tim44 serves as a scaffold for precise positioning of mtHsp70 and its cochaperone Pam18 at the translocon.
Figures



References
-
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. - PubMed
-
- Chacinska A., et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. - PubMed
-
- Craig E. A., Huang P., Aron R., Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006;156:1–21. - PubMed
-
- D'Silva P., Liu Q., Walter W., Craig E. A. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 2004;11:1084–1091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

