Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses
- PMID: 18004277
- PMCID: PMC2132449
- DOI: 10.1038/msb4100188
Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses
Abstract
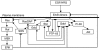
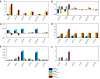
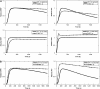
Deregulation of ErbB signaling plays a key role in the progression of multiple human cancers. To help understand ErbB signaling quantitatively, in this work we combine traditional experiments with computational modeling, building a model that describes how stimulation of all four ErbB receptors with epidermal growth factor (EGF) and heregulin (HRG) leads to activation of two critical downstream proteins, extracellular-signal-regulated kinase (ERK) and Akt. Model analysis and experimental validation show that (i) ErbB2 overexpression, which occurs in approximately 25% of all breast cancers, transforms transient EGF-induced signaling into sustained signaling, (ii) HRG-induced ERK activity is much more robust to the ERK cascade inhibitor U0126 than EGF-induced ERK activity, and (iii) phosphoinositol-3 kinase is a major regulator of post-peak but not pre-peak EGF-induced ERK activity. Sensitivity analysis leads to the hypothesis that ERK activation is robust to parameter perturbation at high ligand doses, while Akt activation is not.
Figures



References
-
- Adams GP, Weiner LM (2005) Monoclonal antibody therapy of cancer. Nat Biotechnol 23: 1147–1157 - PubMed
-
- Baulida J, Kraus MH, Alimandi M, Di Fiore PP, Carpenter G (1996) All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem 271: 5251–5257 - PubMed
-
- Berger MB, Mendrola JM, Lemmon MA (2004) ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett 569: 332–336 - PubMed
-
- Blinov ML, Faeder JR, Goldstein B, Hlavacek WS (2006) A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. Biosystems 83: 136–151 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

