A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence
- PMID: 18005689
- PMCID: PMC2083280
- DOI: 10.1016/j.chom.2007.03.001
A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence
Abstract
Staphylococcus aureus, a bacterium responsible for tremendous morbidity and mortality, exists as a harmless commensal in approximately 25% of humans. Identifying the molecular machinery activated upon infection is central to understanding staphylococcal pathogenesis. We describe the heme sensor system (HssRS) that responds to heme exposure and activates expression of the heme-regulated transporter (HrtAB). Inactivation of the Hss or Hrt systems leads to increased virulence in a vertebrate infection model, a phenotype that is associated with an inhibited innate immune response. We suggest that the coordinated activity of Hss and Hrt allows S. aureus to sense internal host tissues, resulting in tempered virulence to avoid excessive host tissue damage. Further, genomic analyses have identified orthologous Hss and Hrt systems in Bacillus anthracis, Listeria monocytogenes, and Enterococcus faecalis, suggesting a conserved regulatory system by which Gram-positive pathogens sense heme as a molecular marker of internal host tissue and modulate virulence.
Figures
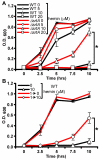
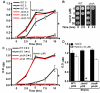



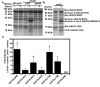

Comment in
-
Did bacterial sensing of host environments evolve from sensing within microbial communities?Cell Host Microbe. 2007 Apr 19;1(2):85-7. doi: 10.1016/j.chom.2007.04.002. Cell Host Microbe. 2007. PMID: 18005684
References
-
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. - PubMed
-
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2005 - PubMed
-
- Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006;9:143–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

