Eelgrass meadows in the California Channel Islands and adjacent coast reveal a mosaic of two species, evidence for introgression and variable clonality
- PMID: 18006507
- PMCID: PMC2701847
- DOI: 10.1093/aob/mcm288
Eelgrass meadows in the California Channel Islands and adjacent coast reveal a mosaic of two species, evidence for introgression and variable clonality
Abstract
Background and aims: Seagrasses are important facilitator species in shallow, soft-bottom marine environments worldwide and, in many places, are threatened by coastal development and eutrophication. One narrow-leaved species (Zostera marina) and one wide-leaved species, variously designated as Z. marina, Z. pacifica or Z. asiatica, are found off the California Channel Islands and adjacent California-Mexico coast. The aim of the present study was to confirm species identification genetically and to link patterns of genetic diversity, connectivity and hybridization among and within the populations with historical sea levels (Ice Age) or the contemporary environment.
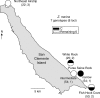
Methods: Samples (n = 11-100) were collected from 28 sites off five California Channel Islands and six sites off the adjacent coast of southern California and Baja California, Mexico. DNA polymorphisms of the rDNA-ITS (internal transcribed spacer) cistron (nuclear), the matK intron (chloroplast) and nine microsatellite loci (nuclear) were examined in a population genetic and phylogeographic context.
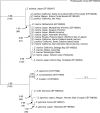
Key results: All wide-leaved individuals were Z. pacifica, whereas narrow-leaved forms were Z. marina. Microsatellite genotypes were consistent with hybridization between the two species in three populations. The present distribution of Z. pacifica follows a glacial age land mass rather than present oceanographic regimes, but no link was observed between the present distribution of Z. marina and past or present environments. Island populations of Z. marina often were clonal and characterized by low genotypic diversity compared with populations along the Baja California coast. The high level of clonal connectivity around Santa Catalina Island indicated the importance of dispersal and subsequent re-establishment of vegetative fragments.
Conclusions: The pristine environmental conditions of offshore islands do not guarantee maximum genetic diversity. Future restoration and transplantation efforts of seagrasses must recognize cryptic species and consider the degree of both genetic and genotypic variation in candidate donor populations.
Figures






References
-
- Alberto F, Arnaud-Haond S, Duarte CM, Serrão EA. Genetic diversity of a clonal angiosperm near its range limit: the case of Cymodocea nodosa at the Canary Islands. Marine Ecology Progress Series. 2006;309:117–129.
-
- Alstatt J. Restoration of a historic eelgrass (Zostera marina) bed at Frenchy's Cove, Anacapa Island. In: Garcelon DK, Schwemm CA, editors. Proceedings of the Sixth California Islands Symposium. Arcata: Institute for Wildlife Studies; 2003. pp. 397–404.
-
- Amos W. The hidden value of missing genotypes. Molecular Biology and Evolution. 2006;23:1995–1996. - PubMed
-
- Anonymous. Southern California eelgrass mitigation policy. Southwest Regional Office, National Marine Fisheries Service, NOAA; 1991.
-
- Anonymous. Understanding marine biodiversity. Washington, DC: National Research Council, National Academy Press; 1995.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

