In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans
- PMID: 18007599
- PMCID: PMC2140100
- DOI: 10.1038/sj.emboj.7601910
In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans
Abstract
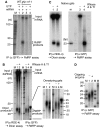
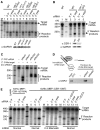
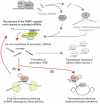
In the RNA interference (RNAi) pathway, small interfering RNAs (siRNAs) play important roles as intermediates. Primary siRNAs are produced from trigger dsRNAs by an RNaseIII-related enzyme called Dicer; in some organisms, secondary siRNAs are also produced by processes involving RNA-dependent RNA polymerases (RdRPs), which act on target mRNAs. Using a cell-free assay system prepared from Caenorhabditis elegans, we analyzed the production and activity of secondary siRNAs. In this cell-free system, RdRP activity acts on mRNA-derived templates to produce small RNAs. The RRF-1 complex is predominantly responsible for the RdRP activity, and synthesizes secondary-type siRNA molecules in a Dicer-independent manner. Notably, secondary-type siRNAs induce a prominent Slicer activity to cleave target mRNAs far more effectively than primary-type siRNAs. An Argonaute protein, CSR-1, is responsible for the Slicer activity induced by secondary-type siRNAs. Secondary rather than primary siRNAs may play a major role in the destabilization of target transcripts during RNAi in C. elegans.
Figures





References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 - PubMed
-
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC (2005) A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol 15: 378–383 - PubMed
-
- Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169 - PubMed
-
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 - PubMed
-
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

