Selective Raf inhibition in cancer therapy
- PMID: 18020980
- PMCID: PMC2720036
- DOI: 10.1517/14728222.11.12.1587
Selective Raf inhibition in cancer therapy
Erratum in
- Expert Opin Ther Targets. 2009 Sep;13(9):1135
Abstract
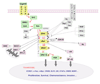
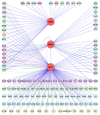
Over the past 5 years, the Raf kinase family has emerged as a promising target for protein-directed cancer therapy development. The goal of this review is to first provide a concise summary of the data validating Raf proteins as high-interest therapeutic targets. The authors then outline the mode of action of Raf kinases, emphasizing how Raf activities and protein interactions suggest specific approaches to inhibiting Raf. The authors then summarize the set of drugs, antisense reagents and antibodies available or in development for therapeutically targeting Raf or Raf-related proteins, as well as existing strategies combining these and other therapeutic agents. Finally, the authors discuss recent results from systems biology analyses that have the potential to increasingly guide the intelligent selection of combination therapies involving Raf-targeting agents and other therapeutics.
Figures

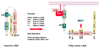

References
-
- Yamanaka Y. The immunohistochemical expressions of epidermal growth factors, epidermal growth factor receptors and c-erbB-2 oncoprotein in human pancreatic cancer. Nippon Ika Daigaku Zasshi. 1992;59(1):51–61. - PubMed
-
- Burtness B. Her signaling in pancreatic cancer. Expert Opin Biol Ther. 2007;7(6):823–829. - PubMed
-
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. - PubMed
-
- Astsaturov I, Cohen RB, Harari P. Targeting epidermal growth factor receptor signaling in the treatment of head and neck cancer. Expert Rev Anticancer Ther. 2006;6(9):1179–1193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
