Experimental-confirmation and functional-annotation of predicted proteins in the chicken genome
- PMID: 18021451
- PMCID: PMC2204016
- DOI: 10.1186/1471-2164-8-425
Experimental-confirmation and functional-annotation of predicted proteins in the chicken genome
Abstract
Background: The chicken genome was sequenced because of its phylogenetic position as a non-mammalian vertebrate, its use as a biomedical model especially to study embryology and development, its role as a source of human disease organisms and its importance as the major source of animal derived food protein. However, genomic sequence data is, in itself, of limited value; generally it is not equivalent to understanding biological function. The benefit of having a genome sequence is that it provides a basis for functional genomics. However, the sequence data currently available is poorly structurally and functionally annotated and many genes do not have standard nomenclature assigned.
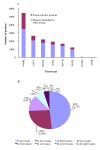
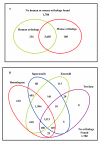
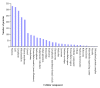
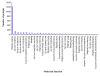
Results: We analysed eight chicken tissues and improved the chicken genome structural annotation by providing experimental support for the in vivo expression of 7,809 computationally predicted proteins, including 30 chicken proteins that were only electronically predicted or hypothetical translations in human. To improve functional annotation (based on Gene Ontology), we mapped these identified proteins to their human and mouse orthologs and used this orthology to transfer Gene Ontology (GO) functional annotations to the chicken proteins. The 8,213 orthology-based GO annotations that we produced represent an 8% increase in currently available chicken GO annotations. Orthologous chicken products were also assigned standardized nomenclature based on current chicken nomenclature guidelines.
Conclusion: We demonstrate the utility of high-throughput expression proteomics for rapid experimental structural annotation of a newly sequenced eukaryote genome. These experimentally-supported predicted proteins were further annotated by assigning the proteins with standardized nomenclature and functional annotation. This method is widely applicable to a diverse range of species. Moreover, information from one genome can be used to improve the annotation of other genomes and inform gene prediction algorithms.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

