Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR
- PMID: 18022369
- PMCID: PMC2170882
- DOI: 10.1016/j.cell.2007.09.039
Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR
Abstract
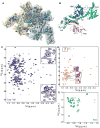
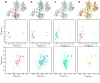
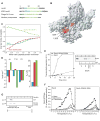
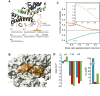
Recognition of signal sequences by cognate receptors controls the entry of virtually all proteins to export pathways. Despite its importance, this process remains poorly understood. Here, we present the solution structure of a signal peptide bound to SecA, the 204 kDa ATPase motor of the Sec translocase. Upon encounter, the signal peptide forms an alpha-helix that inserts into a flexible and elongated groove in SecA. The mode of binding is bimodal, with both hydrophobic and electrostatic interactions mediating recognition. The same groove is used by SecA to recognize a diverse set of signal sequences. Impairment of the signal-peptide binding to SecA results in significant translocation defects. The C-terminal tail of SecA occludes the groove and inhibits signal-peptide binding, but autoinhibition is relieved by the SecB chaperone. Finally, it is shown that SecA interconverts between two conformations in solution, suggesting a simple mechanism for polypeptide translocation.
Figures

References
-
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. - PubMed
-
- Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. - PubMed
-
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Bio Crystallogr. 1998;54:905–921. - PubMed
-
- Chou YT, Gierasch LM. The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA. J Biol Chem. 2005;280:32753–32760. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

