Nuclear receptors: decoding metabolic disease
- PMID: 18023286
- PMCID: PMC2254310
- DOI: 10.1016/j.febslet.2007.11.016
Nuclear receptors: decoding metabolic disease
Abstract
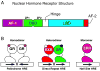
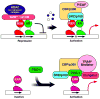
Nuclear receptors (NR) are a superfamily of ligand-activated transcription factors that regulate development, reproduction, and metabolism of lipids, drugs and energy. The importance of this family of proteins in metabolic disease is exemplified by NR ligands used in the clinic or under exploratory development for the treatment of diabetes mellitus, dyslipidemia, hypercholesterolemia, or other metabolic abnormalities. Genetic studies in humans and rodents support the notion that NRs control a wide variety of metabolic processes by regulating the expression of genes encoding key enzymes, transporters and other proteins involved in metabolic homeostasis. Current knowledge of complex NR metabolic networks is summarized here.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

