Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation
- PMID: 18024008
- PMCID: PMC2197162
- DOI: 10.1016/j.sbi.2007.09.011
Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation
Abstract
In recent years, there has been a surge in the number of studies exploring the relationship between proteins' equilibrium dynamics and structural changes involved in function. An emerging concept, supported by both theory and experiments, is that under native state conditions proteins have an intrinsic ability to sample conformations that meet functional requirements. A typical example is the ability of enzymes to sample open and closed forms, irrespective of substrate, succeeded by the stabilization of one form (usually closed) upon substrate binding. This ability is structure-encoded, and plays a key role in facilitating allosteric regulation, which suggests complementing the sequence-encodes-structure paradigm of protein science by structure-encodes-dynamics-encodes-function. The emerging connection implies an evolutionary role in selecting/conserving structures based on their ability to achieve functional dynamics, and in turn, selecting sequences that fold into such 'apt' structures.
Figures
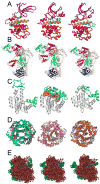
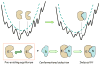
References
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. This is an extensive review of experimental and theoretical evidence supporting the interplay between conformational dynamics of enzymes and their catalytic activities, with focus on two enzymes, dehydrofolate reductase and liver alcohol dehydrogenase. A network of coupled motions involving both fast thermal vibrations and low frequency modes is pointed out to modulate the interactions between enzyme, substrate and cofactor at the active site, facilitating the hydride transfer reaction. - PubMed
-
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. This is an insightful review of the progress made in the last forty years in understanding the allosteric mechanisms of signal tranduction. The basic concepts underlying the MWC model are summarized, such as the pre-disposition of symmetric oligomeric structures to undergo cooperative changes and the occurrence of reversible transitions, or spontaneous ‘switches’ between discrete conformations, accessible in the absence of ligand. The importance of binding ligands at strategic locations such as interfaces between subunits or along symmetry axes is emphasized. - PubMed
-
- Tama F, Brooks CL. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu Rev Biophys Biomol Struct. 2006;35:115–133. An excellent review where the authors draw attention to the utility of multi-resolution methods based on elastic network models for understanding the functional dynamics of biological systems. They highlight the role of shape and form in the defining robust modes of motions. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

