Penicillin-binding proteins and cell wall composition in beta-lactam-sensitive and -resistant strains of Staphylococcus sciuri
- PMID: 18024515
- PMCID: PMC2223715
- DOI: 10.1128/JB.01549-07
Penicillin-binding proteins and cell wall composition in beta-lactam-sensitive and -resistant strains of Staphylococcus sciuri
Abstract
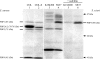
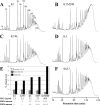
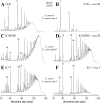
A close homologue of the acquired Staphylococcus aureus mecA gene is present as a native gene in Staphylococcus sciuri. We determined the patterns of penicillin-binding proteins (PBPs) and the peptidoglycan compositions of several S. sciuri strains to explore the functions of this mecA homologue, named pbpD, in its native S. sciuri environment. The protein product of pbpD was identified as PBP4 with a molecular mass of 84 kDa, one of the six PBPs present in representatives of each of three subspecies of S. sciuri examined. PBP4 had a low affinity for nafcillin, reacted with a monoclonal antibody raised against S. aureus PBP2A, and was greatly overproduced in oxacillin-resistant clinical isolate S. sciuri SS37 and to a lesser extent in resistant laboratory mutant K1M200. An additional PBP inducible by oxacillin and corresponding to S. aureus PBP2A was identified in another oxacillin-resistant clinical isolate, S. sciuri K3, which harbors an S. aureus copy of mecA. Oxacillin resistance depended on the overtranscribed S. sciuri pbpD gene in strains SS37 and K1M200, while the resistance of strain K3 depended on the S. aureus copy of mecA. Our data provide evidence that both S. aureus mecA and S. sciuri pbpD can function as resistance determinants in either an S. aureus or an S. sciuri background and that the protein products of these genes, S. aureus PBP2A and S. sciuri PBP4, can participate in the biosynthesis of peptidoglycan, the muropeptide composition of which depends on the bacterium "hosting" the resistance gene.
Figures



References
-
- Couto, I., H. de Lencastre, E. Severina, W. Kloos, J. A. Webster, R. J. Hubner, I. S. Sanches, and A. Tomasz. 1996. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2377-391. - PubMed
-
- de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 26711248-11254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

