Structure, evolution, and biology of the MUC4 mucin
- PMID: 18024835
- PMCID: PMC2835492
- DOI: 10.1096/fj.07-9673rev
Structure, evolution, and biology of the MUC4 mucin
Abstract
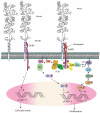
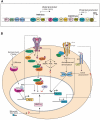
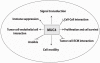
Mucins are high-molecular-weight glycoproteins and are implicated in diverse biological functions. MUC4, a member of transmembrane mucin family, is expressed in airway epithelial cells and body fluids like saliva, tear film, ear fluid, and breast milk. In addition to its normal expression, an aberrant expression of MUC4 has been reported in a variety of carcinomas. Among various potential domains of MUC4, epidermal growth factor (EGF) -like domains are hypothesized to interact with and activate the ErbB2 receptors, suggesting an intramembrane-growth factor function for MUC4. The heavily glycosylated tandem repeat domain provides the structural rigidity to the extended extracellular region. MUC4, by virtue of its extended structure, serves as a barrier for some cell-cell and cell-extracellular matrix interactions and as a potential reservoir for certain growth factors. An intricate relationship between MUC4 and growth factor signaling is also reflected in the transcriptional regulation of MUC4. The MUC4 promoter has binding sites for different transcription factors, which are responsible for the regulation of its expression in different tissues. The interferon-gamma, retinoic acid, and transforming growth factor-beta signaling pathways regulate MUC4 expression in a partially interdependent manner. Taken together, all of these features of MUC4 strongly support its role as a potential candidate for diagnostic and therapeutic applications in cancer and other diseases.
Figures



References
-
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer. 2004;4:45–60. - PubMed
-
- Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front. Biosci. 2001;6:D1192–1206. - PubMed
-
- Satoh S, Hinoda Y, Hayashi T, Burdick MD, Imai K, Hollingsworth MA. Enhancement of metastatic properties of pancreatic cancer cells by MUC1 gene encoding an anti-adhesion molecule. Int. J. Cancer. 2000;88:507–518. - PubMed
-
- Strous GJ, Dekker J. Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol. 1992;27:57–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

