Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers
- PMID: 18025472
- PMCID: PMC2141882
- DOI: 10.1073/pnas.0706114104
Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers
Abstract
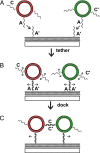
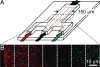
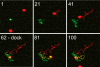
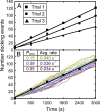
Membrane-membrane recognition and binding are crucial in many biological processes. We report an approach to studying the dynamics of such reactions by using DNA-tethered vesicles as a general scaffold for displaying membrane components. This system was used to characterize the docking reaction between two populations of tethered vesicles that display complementary DNA. Deposition of vesicles onto a supported lipid bilayer was performed by using a microfluidic device to prevent mixing of the vesicles in bulk during sample preparation. Once tethered onto the surface, vesicles mixed via two-dimensional diffusion. DNA-mediated docking of two reacting vesicles results in their colocalization after collision and their subsequent tandem motion. Individual docking events and population kinetics were observed via epifluorescence microscopy. A lattice-diffusion simulation was implemented to extract from experimental data the probability, P(dock), that a collision leads to docking. For individual vesicles displaying small numbers of docking DNA, P(dock) shows a first-order relationship with copy number as well as a strong dependence on the DNA sequence. Both trends are explained by a model that includes both tethered vesicle diffusion on the supported bilayer and docking DNA diffusion over each vesicle's surface. These results provide the basis for the application of tethered vesicles to study other membrane reactions including protein-mediated docking and fusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

