Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY
- PMID: 18026095
- PMCID: PMC2759348
- DOI: 10.1038/nchembio.2007.40
Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY
Abstract
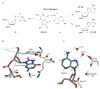
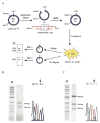
Escherichia coli MutY has an important role in preventing mutations associated with the oxidative lesion 7,8-dihydro-8-oxo-2'-deoxyguanosine (OG) in DNA by excising adenines from OG.A mismatches as the first step of base excision repair. To determine the importance of specific steps in the base pair recognition and base removal process of MutY, we have evaluated the effects of modifications of the OG.A substrate on the kinetics of base removal, mismatch affinity and repair to G-C in an E. coli-based assay. Notably, adenine modification was tolerated in the cellular assay, whereas modification of OG resulted in minimal cellular repair. High affinity for the mismatch and efficient base removal required the presence of OG. Taken together, these results suggest that the presence of OG is a critical feature that is necessary for MutY to locate OG.A mismatches and select the appropriate adenines for excision to initiate repair in vivo before replication.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

