A de novo designed protein protein interface
- PMID: 18029425
- PMCID: PMC2222823
- DOI: 10.1110/ps.073125207
A de novo designed protein protein interface
Abstract
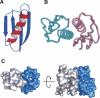
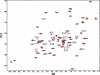
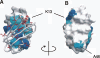
As an approach to both explore the physical/chemical parameters that drive molecular self-assembly and to generate novel protein oligomers, we have developed a procedure to generate protein dimers from monomeric proteins using computational protein docking and amino acid sequence design. A fast Fourier transform-based docking algorithm was used to generate a model for a dimeric version of the 56-amino-acid beta1 domain of streptococcal protein G. Computational amino acid sequence design of 24 residues at the dimer interface resulted in a heterodimer comprised of 12-fold and eightfold variants of the wild-type protein. The designed proteins were expressed, purified, and characterized using analytical ultracentrifugation and heteronuclear NMR techniques. Although the measured dissociation constant was modest ( approximately 300 microM), 2D-[(1)H,(15)N]-HSQC NMR spectra of one of the designed proteins in the absence and presence of its binding partner showed clear evidence of specific dimer formation.
Figures
References
-
- Chevalier B.S., Kortemme, T., Chadsey, M.S., Baker, D., Monnat, R.J., and Stoddard, B.L. 2002. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10: 895–905. - PubMed
-
- Clackson T. and Wells, J.A. 1995. A hot-spot of binding-energy in a hormone–receptor interface. Science 267: 383–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

