The amazing diversity of cap-independent translation elements in the 3'-untranslated regions of plant viral RNAs
- PMID: 18031280
- PMCID: PMC3081161
- DOI: 10.1042/BST0351629
The amazing diversity of cap-independent translation elements in the 3'-untranslated regions of plant viral RNAs
Abstract
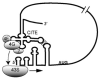
Many plant viral RNAs lack the 5'-cap structure that is required on all host mRNAs for interacting with essential translation initiation factors. Instead, uncapped viral RNAs take over the host translation machinery by harbouring sequences that functionally replace the 5'-cap. Recent reports reveal at least eight different classes of CITE (cap-independent translation element) located in the 3'-UTRs (untranslated regions) of various viruses. We describe how the structure and behaviour of each class of element differs from the other classes, suggesting that they recruit translation factors and, ultimately, the ribosome by diverse mechanisms. These results greatly expand our understanding of ways in which mRNAs can recruit ribosomes, and they provide insight into the regulation of virus gene expression.
Figures

References
-
- Flint SJ, Enquist LW, Racaniello VR, Skalka AM. Principles of Virology. ASM Press; Washington, DC: 2004.
-
- Gallie DR. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey J, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 747–774.
-
- Pestova TV, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey J, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 87–128.
-
- Browning KS. Biochem. Soc. Trans. 2004;32:589–591. - PubMed
-
- Jan E. Virus Res. 2006;119:16–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

