Transcription of hepatitis delta virus RNA by RNA polymerase II
- PMID: 18032511
- PMCID: PMC2224410
- DOI: 10.1128/JVI.01758-07
Transcription of hepatitis delta virus RNA by RNA polymerase II
Abstract
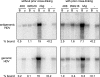
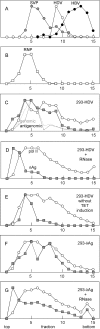
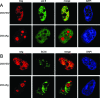
Previous studies have indicated that the replication of the RNA genome of hepatitis delta virus (HDV) involves redirection of RNA polymerase II (Pol II), a host enzyme that normally uses DNA as a template. However, there has been some controversy about whether in one part of this HDV RNA transcription, a polymerase other than Pol II is involved. The present study applied a recently described cell system (293-HDV) of tetracycline-inducible HDV RNA replication to provide new data regarding the involvement of host polymerases in HDV transcription. The data generated with a nuclear run-on assay demonstrated that synthesis not only of genomic RNA but also of its complement, the antigenome, could be inhibited by low concentrations of amanitin specific for Pol II transcription. Subsequent studies used immunoprecipitation and rate-zonal sedimentation of nuclear extracts together with double immunostaining of 293-HDV cells, in order to examine the associations between Pol II and HDV RNAs, as well as the small delta antigen, an HDV-encoded protein known to be essential for replication. Findings include evidence that HDV replication is somehow able to direct the available delta antigen to sites in the nucleoplasm, almost exclusively colocalized with Pol II in what others have described as transcription factories.
Figures



References
-
- Chang, J., X. Nie, S. Gudima, and J. Taylor. 2006. Replication of the hepatitis delta virus genome, p. 195-212. In K. L. Hefferon (ed.), Recent advances in RNA virus replication. Transworld Research Network, Kerala, India.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

