Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates
- PMID: 18032721
- PMCID: PMC2134780
- DOI: 10.1101/gr.7020108
Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates
Abstract
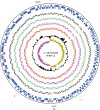
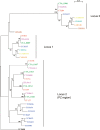
Chlamydia trachomatis is the most common cause of sexually transmitted infections in the UK, a statistic that is also reflected globally. There are three biovariants of C. trachomatis: trachoma (serotypes A-C) and two sexually transmitted pathovars; serotypes D-K and lymphogranuloma venereum (LGV). Trachoma isolates and the sexually transmitted serotypes D-K are noninvasive, whereas the LGV strains are invasive, causing a disseminating infection of the local draining lymph nodes. Genome sequences are available for single isolates from the trachoma (serotype A) and sexually transmitted (serotype D) biotypes. We sequenced two isolates from the remaining biotype, LGV, a long-term laboratory passaged strain and the recent "epidemic" LGV isolate-causing proctitis. Although the genome of the LGV strain shows no additional genes that could account for the differences in disease outcome, we found evidence of functional gene loss and identified regions of heightened sequence variation that have previously been shown to be important sites for interstrain recombination. We have used new sequencing technologies to show that the recent clinical LGV isolate causing proctitis is unlikely to be a newly emerged strain but is most probably an old strain with relatively new clinical manifestations.
Figures



References
-
- Azuma Y., Hirakawa H., Yamashita A., Cai Y., Rahman M.A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Hirakawa H., Yamashita A., Cai Y., Rahman M.A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Yamashita A., Cai Y., Rahman M.A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Cai Y., Rahman M.A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Rahman M.A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Mitaku S., Toh H., Goto S., Murakami T., Toh H., Goto S., Murakami T., Goto S., Murakami T., Murakami T., et al. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 2006;13:15–23. - PubMed
-
- Bannantine J.P., Rockey D.D., Hackstadt T., Rockey D.D., Hackstadt T., Hackstadt T. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 1998a;28:1017–1026. - PubMed
-
- Bannantine J.P., Stamm W.E., Suchland R.J., Rockey D.D., Stamm W.E., Suchland R.J., Rockey D.D., Suchland R.J., Rockey D.D., Rockey D.D. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 1998b;66:6017–6021. - PMC - PubMed
-
- Bannantine J.P., Griffiths R.S., Viratyosin W., Brown W.J., Rockey D.D., Griffiths R.S., Viratyosin W., Brown W.J., Rockey D.D., Viratyosin W., Brown W.J., Rockey D.D., Brown W.J., Rockey D.D., Rockey D.D. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2000;2:35–47. - PubMed
-
- Behets F.M., Andriamiadana J., Randrianasolo D., Randriamanga R., Rasamilalao D., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Andriamiadana J., Randrianasolo D., Randriamanga R., Rasamilalao D., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Randrianasolo D., Randriamanga R., Rasamilalao D., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Randriamanga R., Rasamilalao D., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Rasamilalao D., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Chen C.Y., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Weiss J.B., Morse S.A., Dallabetta G., Cohen M.S., Morse S.A., Dallabetta G., Cohen M.S., Dallabetta G., Cohen M.S., Cohen M.S. Chancroid, primary syphilis, genital herpes, and lymphogranuloma venereum in Antananarivo, Madagascar. J. Infect. Dis. 1999;180:1382–1385. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
