Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development
- PMID: 18034876
- PMCID: PMC2220006
- DOI: 10.1186/1471-2164-8-429
Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development
Abstract
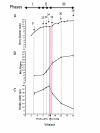
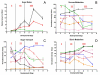
Background: Grape berry development is a dynamic process that involves a complex series of molecular genetic and biochemical changes divided into three major phases. During initial berry growth (Phase I), berry size increases along a sigmoidal growth curve due to cell division and subsequent cell expansion, and organic acids (mainly malate and tartrate), tannins, and hydroxycinnamates accumulate to peak levels. The second major phase (Phase II) is defined as a lag phase in which cell expansion ceases and sugars begin to accumulate. Véraison (the onset of ripening) marks the beginning of the third major phase (Phase III) in which berries undergo a second period of sigmoidal growth due to additional mesocarp cell expansion, accumulation of anthocyanin pigments for berry color, accumulation of volatile compounds for aroma, softening, peak accumulation of sugars (mainly glucose and fructose), and a decline in organic acid accumulation. In order to understand the transcriptional network responsible for controlling berry development, mRNA expression profiling was conducted on berries of V. vinifera Cabernet Sauvignon using the Affymetrix GeneChip Vitis oligonucleotide microarray ver. 1.0 spanning seven stages of berry development from small pea size berries (E-L stages 31 to 33 as defined by the modified E-L system), through véraison (E-L stages 34 and 35), to mature berries (E-L stages 36 and 38). Selected metabolites were profiled in parallel with mRNA expression profiling to understand the effect of transcriptional regulatory processes on specific metabolite production that ultimately influence the organoleptic properties of wine.
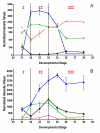
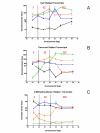
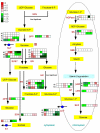
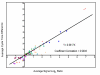
Results: Over the course of berry development whole fruit tissues were found to express an average of 74.5% of probes represented on the Vitis microarray, which has 14,470 Unigenes. Approximately 60% of the expressed transcripts were differentially expressed between at least two out of the seven stages of berry development (28% of transcripts, 4,151 Unigenes, had pronounced (> or =2 fold) differences in mRNA expression) illustrating the dynamic nature of the developmental process. The subset of 4,151 Unigenes was split into twenty well-correlated expression profiles. Expression profile patterns included those with declining or increasing mRNA expression over the course of berry development as well as transient peak or trough patterns across various developmental stages as defined by the modified E-L system. These detailed surveys revealed the expression patterns for genes that play key functional roles in phytohormone biosynthesis and response, calcium sequestration, transport and signaling, cell wall metabolism mediating expansion, ripening, and softening, flavonoid metabolism and transport, organic and amino acid metabolism, hexose sugar and triose phosphate metabolism and transport, starch metabolism, photosynthesis, circadian cycles and pathogen resistance. In particular, mRNA expression patterns of transcription factors, abscisic acid (ABA) biosynthesis, and calcium signaling genes identified candidate factors likely to participate in the progression of key developmental events such as véraison and potential candidate genes associated with such processes as auxin partitioning within berry cells, aroma compound production, and pathway regulation and sequestration of flavonoid compounds. Finally, analysis of sugar metabolism gene expression patterns indicated the existence of an alternative pathway for glucose and triose phosphate production that is invoked from véraison to mature berries.
Conclusion: These results reveal the first high-resolution picture of the transcriptome dynamics that occur during seven stages of grape berry development. This work also establishes an extensive catalog of gene expression patterns for future investigations aimed at the dissection of the transcriptional regulatory hierarchies that govern berry development in a widely grown cultivar of wine grape. More importantly, this analysis identified a set of previously unknown genes potentially involved in critical steps associated with fruit development that can now be subjected to functional testing.
Figures



References
-
- FAOSTAT database (http://faostat.fao.org) Food and Agriculture Organization of the United Nations; 2007.
-
- Wine Institute http://www.wineinstitute.org/
-
- Wollin SD, Jones PJ. Alcohol, red wine and cardiovascular disease. J Nutr. 2001;131:1401–1404. - PubMed
-
- Bradamante S, Barenghi L, Villa A. Cardiovascular protective effects of resveratrol. Cardiovasc Drug Rev. 2004;22:169–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

