MADS-complexes regulate transcriptome dynamics during pollen maturation
- PMID: 18034896
- PMCID: PMC2258202
- DOI: 10.1186/gb-2007-8-11-r249
MADS-complexes regulate transcriptome dynamics during pollen maturation
Abstract
Background: Differentiation processes are responsible for the diversity and functional specialization of the cell types that compose an organism. The outcome of these processes can be studied at molecular, physiologic, and biochemical levels by comparing different cell types, but the complexity and dynamics of the regulatory processes that specify the differentiation are largely unexplored.
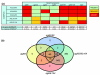
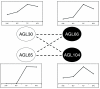
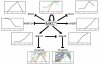
Results: Here we identified the pollen-specific MIKC* class of MADS-domain transcription factors as major regulators of transcriptome dynamics during male reproductive cell development in Arabidopsis thaliana. Pollen transcript profiling of mutants deficient in different MIKC* protein complexes revealed that they control a transcriptional switch that directs pollen maturation and that is essential for pollen competitive ability. We resolved the functional redundancy among the MIKC* proteins and uncovered part of the underlying network by identifying the non-MIKC* MADS-box genes AGL18 and AGL29 as downstream regulators of a subset of the MIKC* MADS-controlled genes.
Conclusion: Our results provide a first, unique, and compelling insight into the complexity of a transcription factor network that directs cellular differentiation during pollen maturation, a process that is essential for male reproductive fitness in flowering plants.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

