Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III
- PMID: 18039033
- PMCID: PMC2082468
- DOI: 10.1371/journal.pgen.0030212
Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III
Abstract
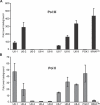
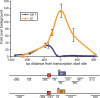
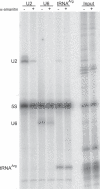
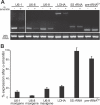
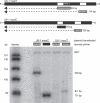
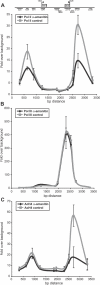
Recent genomic data indicate that RNA polymerase II (Pol II) function extends beyond conventional transcription of primarily protein-coding genes. Among the five snRNAs required for pre-mRNA splicing, only the U6 snRNA is synthesized by RNA polymerase III (Pol III). Here we address the question of how Pol II coordinates the expression of spliceosome components, including U6. We used chromatin immunoprecipitation (ChIP) and high-resolution mapping by PCR to localize both Pol II and Pol III to snRNA gene regions. We report the surprising finding that Pol II is highly concentrated approximately 300 bp upstream of all five active human U6 genes in vivo. The U6 snRNA, an essential component of the spliceosome, is synthesized by Pol III, whereas all other spliceosomal snRNAs are Pol II transcripts. Accordingly, U6 transcripts were terminated in a Pol III-specific manner, and Pol III localized to the transcribed gene regions. However, synthesis of both U6 and U2 snRNAs was alpha-amanitin-sensitive, indicating a requirement for Pol II activity in the expression of both snRNAs. Moreover, both Pol II and histone tail acetylation marks were lost from U6 promoters upon alpha-amanitin treatment. The results indicate that Pol II is concentrated at specific genomic regions from which it can regulate Pol III activity by a general mechanism. Consequently, Pol II coordinates expression of all RNA and protein components of the spliceosome.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. 1988;334:213–218. - PubMed
-
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

