Notch signaling in leukemia
- PMID: 18039126
- PMCID: PMC5934586
- DOI: 10.1146/annurev.pathmechdis.3.121806.154300
Notch signaling in leukemia
Abstract
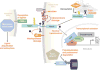
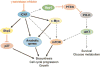
Recent discoveries indicate that gain-of-function mutations in the Notch1 receptor are very common in human T cell acute lymphoblastic leukemia/lymphoma. This review discusses what these mutations have taught us about normal and pathophysiologic Notch1 signaling, and how these insights may lead to new targeted therapies for patients with this aggressive form of cancer.
Figures
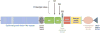

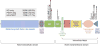






References
-
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. - PubMed
-
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. Describes the discovery of Notch1 through analysis of the t(7;9) found in T-ALL and provides the first suggestion that truncation of Notch1 via proteolysis might play a role in its activation. - PubMed
-
- Aster J, Pear W, Hasserjian R, Erba H, Davi F, et al. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harb Symp Quant Biol. 1994;59:125–36. - PubMed
-
- Das I, Craig C, Funahashi Y, Jung KM, Kim TW, et al. Notch oncoproteins depend on γ-secretase/presenilin activity for processing and function. J Biol Chem. 2004;279:30771–80. - PubMed
-
- Palomero T, Barnes KC, Real PJ, Bender JL, Sulis ML, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to γ-secretase inhibitors. Leukemia. 2006;20:1279–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

