Transcription of Satellite III non-coding RNAs is a general stress response in human cells
- PMID: 18039709
- PMCID: PMC2241877
- DOI: 10.1093/nar/gkm1056
Transcription of Satellite III non-coding RNAs is a general stress response in human cells
Abstract
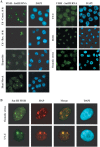
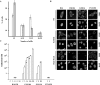
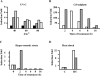
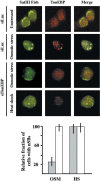
In heat-shocked human cells, heat shock factor 1 activates transcription of tandem arrays of repetitive Satellite III (SatIII) DNA in pericentromeric heterochromatin. Satellite III RNAs remain associated with sites of transcription in nuclear stress bodies (nSBs). Here we use real-time RT-PCR to study the expression of these genomic regions. Transcription is highly asymmetrical and most of the transcripts contain the G-rich strand of the repeat. A low level of G-rich RNAs is detectable in unstressed cells and a 10(4)-fold induction occurs after heat shock. G-rich RNAs are induced by a wide range of stress treatments including heavy metals, UV-C, oxidative and hyper-osmotic stress. Differences exist among stressing agents both for the kinetics and the extent of induction (>100- to 80.000-fold). In all cases, G-rich transcripts are associated with nSBs. On the contrary, C-rich transcripts are almost undetectable in unstressed cells and modestly increase after stress. Production of SatIII RNAs after hyper-osmotic stress depends on the Tonicity Element Binding Protein indicating that activation of the arrays is triggered by different transcription factors. This is the first example of a non-coding RNA whose transcription is controlled by different transcription factors under different growth conditions.
Figures
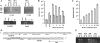


References
-
- Morimoto RI, Sarge KD, Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J. Biol. Chem. 1992;267:21987–21990. - PubMed
-
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. Faseb J. 2001;15:1118–1131. - PubMed
-
- Herr I, Debatin KM. Cellular stress response and apoptosis in cancer therapy. Blood. 2001;98:2603–2614. - PubMed
-
- Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

