Transcription profiling of the stringent response in Escherichia coli
- PMID: 18039766
- PMCID: PMC2223561
- DOI: 10.1128/JB.01092-07
Transcription profiling of the stringent response in Escherichia coli
Abstract
The bacterial stringent response serves as a paradigm for understanding global regulatory processes. It can be triggered by nutrient downshifts or starvation and is characterized by a rapid RelA-dependent increase in the alarmone (p)ppGpp. One hallmark of the response is the switch from maximum-growth-promoting to biosynthesis-related gene expression. However, the global transcription patterns accompanying the stringent response in Escherichia coli have not been analyzed comprehensively. Here, we present a time series of gene expression profiles for two serine hydroxymate-treated cultures: (i) MG1655, a wild-type E. coli K-12 strain, and (ii) an isogenic relADelta251 derivative defective in the stringent response. The stringent response in MG1655 develops in a hierarchical manner, ultimately involving almost 500 differentially expressed genes, while the relADelta251 mutant response is both delayed and limited in scope. We show that in addition to the down-regulation of stable RNA-encoding genes, flagellar and chemotaxis gene expression is also under stringent control. Reduced transcription of these systems, as well as metabolic and transporter-encoding genes, constitutes much of the down-regulated expression pattern. Conversely, a significantly larger number of genes are up-regulated. Under the conditions used, induction of amino acid biosynthetic genes is limited to the leader sequences of attenuator-regulated operons. Instead, up-regulated genes with known functions, including both regulators (e.g., rpoE, rpoH, and rpoS) and effectors, are largely involved in stress responses. However, one-half of the up-regulated genes have unknown functions. How these results are correlated with the various effects of (p)ppGpp (in particular, RNA polymerase redistribution) is discussed.
Figures
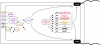
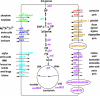
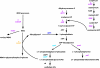
References
-
- Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52613-619. - PubMed
-
- Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117299-310. - PubMed
-
- Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 289-22. - PubMed
-
- Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305689-702. - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305673-688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

