A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila
- PMID: 18039839
- PMCID: PMC2223392
- DOI: 10.1128/MCB.01622-07
A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila
Abstract
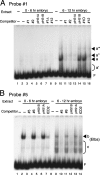
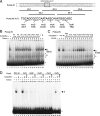
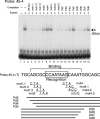
The Fab-7 boundary is required to ensure that the iab-6 and iab-7 cis-regulatory domains in the Drosophila Bithorax complex can function autonomously. Though Fab-7 functions as a boundary from early embryogenesis through to the adult stage, this constitutive boundary activity depends on subelements whose activity is developmentally restricted. In the studies reported here, we have identified a factor, called early boundary activity (Elba), that confers Fab-7 boundary activity during early embryogenesis. The Elba factor binds to a recognition sequence within a Fab-7 subelement that has enhancer-blocking activity during early embryogenesis, but not during mid-embryogenesis or in the adult. We found that the Elba factor is present in early embryos but largely disappears during mid-embryogenesis. We show that mutations in the Elba recognition sequence that eliminate Elba binding in nuclear extracts disrupt the early boundary activity of the Fab-7 subelement. Conversely, we find that early boundary activity can be reconstituted by multimerizing the Elba recognition site.
Figures

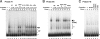



References
-
- Akbari, O. S., A. Bousum, E. Bae, and R. A. Drewell. 2006. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev. Biol. 293294-304. - PubMed
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98387-396. - PubMed
-
- Gaszner, M., and G. Felsenfeld. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7703-713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
