Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii
- PMID: 18039939
- PMCID: PMC2238169
- DOI: 10.1128/EC.00361-07
Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii
Abstract
The highly conserved target of rapamycin (TOR) kinase is a central controller of cell growth in all eukaryotes. TOR exists in two functionally and structurally distinct complexes, termed TOR complex 1 (TORC1) and TORC2. LST8 is a TOR-interacting protein that is present in both TORC1 and TORC2. Here we report the identification and characterization of TOR and LST8 in large protein complexes in the model photosynthetic green alga Chlamydomonas reinhardtii. We demonstrate that Chlamydomonas LST8 is part of a rapamycin-sensitive TOR complex in this green alga. Biochemical fractionation and indirect immunofluorescence microscopy studies indicate that TOR and LST8 exist in high-molecular-mass complexes that associate with microsomal membranes and are particularly abundant in the peri-basal body region in Chlamydomonas cells. A Saccharomyces cerevisiae complementation assay demonstrates that Chlamydomonas LST8 is able to functionally and structurally replace endogenous yeast LST8 and allows us to propose that binding of LST8 to TOR is essential for cell growth.
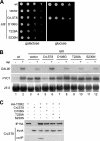
Figures




Similar articles
-
The TOR Signaling Network in the Model Unicellular Green Alga Chlamydomonas reinhardtii.Biomolecules. 2017 Jul 12;7(3):54. doi: 10.3390/biom7030054. Biomolecules. 2017. PMID: 28704927 Free PMC article. Review.
-
Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii.Plant Physiol. 2005 Dec;139(4):1736-49. doi: 10.1104/pp.105.070847. Epub 2005 Nov 18. Plant Physiol. 2005. PMID: 16299168 Free PMC article.
-
Phosphorus Availability Regulates TORC1 Signaling via LST8 in Chlamydomonas.Plant Cell. 2020 Jan;32(1):69-80. doi: 10.1105/tpc.19.00179. Epub 2019 Nov 11. Plant Cell. 2020. PMID: 31712405 Free PMC article.
-
Evolutionary conservation of TORC1 components, TOR, Raptor, and LST8, between rice and yeast.Mol Genet Genomics. 2015 Oct;290(5):2019-30. doi: 10.1007/s00438-015-1056-0. Epub 2015 May 9. Mol Genet Genomics. 2015. PMID: 25956502
-
Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: a review.Biochimie. 2014 May;100:207-18. doi: 10.1016/j.biochi.2013.10.006. Epub 2013 Oct 15. Biochimie. 2014. PMID: 24139906 Review.
Cited by
-
Algal Autophagy Is Necessary for the Regulation of Carbon Metabolism Under Nutrient Deficiency.Front Plant Sci. 2020 Feb 5;11:36. doi: 10.3389/fpls.2020.00036. eCollection 2020. Front Plant Sci. 2020. PMID: 32117375 Free PMC article. Review.
-
TOR signaling in plants: conservation and innovation.Development. 2018 Jul 9;145(13):dev160887. doi: 10.1242/dev.160887. Development. 2018. PMID: 29986898 Free PMC article. Review.
-
Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12526-34. doi: 10.1073/pnas.1302455110. Epub 2013 Jul 12. Proc Natl Acad Sci U S A. 2013. PMID: 23852728 Free PMC article.
-
Autophagy in protists.Autophagy. 2011 Feb;7(2):127-58. doi: 10.4161/auto.7.2.13310. Epub 2011 Feb 1. Autophagy. 2011. PMID: 20962583 Free PMC article. Review.
-
A downstream box fusion allows stable accumulation of a bacterial cellulase in Chlamydomonas reinhardtii chloroplasts.Biotechnol Biofuels. 2018 May 10;11:133. doi: 10.1186/s13068-018-1127-7. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29760775 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

