Acetylcholine receptor gating at extracellular transmembrane domain interface: the cys-loop and M2-M3 linker
- PMID: 18040057
- PMCID: PMC2151658
- DOI: 10.1085/jgp.200709856
Acetylcholine receptor gating at extracellular transmembrane domain interface: the cys-loop and M2-M3 linker
Abstract
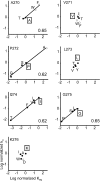
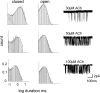
Acetylcholine receptor channel gating is a propagated conformational cascade that links changes in structure and function at the transmitter binding sites in the extracellular domain (ECD) with those at a "gate" in the transmembrane domain (TMD). We used Phi-value analysis to probe the relative timing of the gating motions of alpha-subunit residues located near the ECD-TMD interface. Mutation of four of the seven amino acids in the M2-M3 linker (which connects the pore-lining M2 helix with the M3 helix), including three of the four residues in the core of the linker, changed the diliganded gating equilibrium constant (K(eq)) by up to 10,000-fold (P272 > I274 > A270 > G275). The average Phi-value for the whole linker was approximately 0.64. One interpretation of this result is that the gating motions of the M2-M3 linker are approximately synchronous with those of much of M2 (approximately 0.64), but occur after those of the transmitter binding site region (approximately 0.93) and loops 2 and 7 (approximately 0.77). We also examined mutants of six cys-loop residues (V132, T133, H134, F135, P136, and F137). Mutation of V132, H134, and F135 changed K(eq) by 2800-, 10-, and 18-fold, respectively, and with an average Phi-value of 0.74, similar to those of other cys-loop residues. Even though V132 and I274 are close, the energetic coupling between I and V mutants of these positions was small (< or =0.51 kcal mol(-1)). The M2-M3 linker appears to be the key moving part that couples gating motions at the base of the ECD with those in TMD. These interactions are distributed along an approximately 16-A border and involve about a dozen residues.
Figures
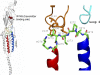




Similar articles
-
Acetylcholine receptor gating at extracellular transmembrane domain interface: the "pre-M1" linker.J Gen Physiol. 2007 Dec;130(6):559-68. doi: 10.1085/jgp.200709857. J Gen Physiol. 2007. PMID: 18040058 Free PMC article.
-
Acetylcholine receptor gating: movement in the alpha-subunit extracellular domain.J Gen Physiol. 2007 Dec;130(6):569-79. doi: 10.1085/jgp.200709858. J Gen Physiol. 2007. PMID: 18040059 Free PMC article.
-
Conformational dynamics of the alphaM3 transmembrane helix during acetylcholine receptor channel gating.Biophys J. 2007 Aug 1;93(3):859-65. doi: 10.1529/biophysj.107.105171. Epub 2007 May 18. Biophys J. 2007. PMID: 17513382 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
-
Gating mechanisms in Cys-loop receptors.Eur Biophys J. 2009 Dec;39(1):37-49. doi: 10.1007/s00249-009-0452-y. Epub 2009 Apr 29. Eur Biophys J. 2009. PMID: 19404635 Review.
Cited by
-
Size matters in activation/inhibition of ligand-gated ion channels.Trends Pharmacol Sci. 2012 Sep;33(9):482-93. doi: 10.1016/j.tips.2012.06.005. Epub 2012 Jul 11. Trends Pharmacol Sci. 2012. PMID: 22789930 Free PMC article. Review.
-
Contributions of conserved residues at the gating interface of glycine receptors.J Biol Chem. 2011 Oct 7;286(40):35129-36. doi: 10.1074/jbc.M111.269027. Epub 2011 Aug 11. J Biol Chem. 2011. PMID: 21835920 Free PMC article.
-
Signal transduction pathways in the pentameric ligand-gated ion channels.PLoS One. 2013 May 8;8(5):e64326. doi: 10.1371/journal.pone.0064326. Print 2013. PLoS One. 2013. PMID: 23667707 Free PMC article.
-
Unmasking coupling between channel gating and ion permeation in the muscle nicotinic receptor.Elife. 2021 Apr 6;10:e66225. doi: 10.7554/eLife.66225. Elife. 2021. PMID: 33821794 Free PMC article.
-
Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions.PLoS Biol. 2013 Nov;11(11):e1001714. doi: 10.1371/journal.pbio.1001714. Epub 2013 Nov 19. PLoS Biol. 2013. PMID: 24260024 Free PMC article.
References
-
- Boehr, D.D., H.J. Dyson, and P.E. Wright. 2006. An NMR perspective on enzyme dynamics. Chem. Rev. 106:3055–3079. - PubMed

