Development of a TaqMan assay for the six major genotypes of hepatitis C virus: comparison with commercial assays
- PMID: 18041021
- PMCID: PMC5653921
- DOI: 10.1002/jmv.21043
Development of a TaqMan assay for the six major genotypes of hepatitis C virus: comparison with commercial assays
Abstract
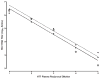
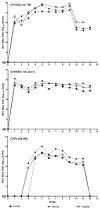
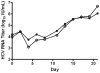
A quantitative real-time PCR assay was developed that detects genomic RNA from reference strains representing the six major genotypes of hepatitis C virus (HCV) with equal sensitivity and accurately measured HCV RNA in JFH1 HCV-infected Huh7.5 cells. The method is indirectly calibrated to the first international (WHO 96/790) HCV standard preparation and has a linear dynamic range of 10(2.6)-10(6.5) IU/ml. In addition, the inter- and intra-assay precision were approximately 3% CV and <2% CV, respectively. Comparison with results obtained by commercially available HCV RNA Nucleic Acid Technology kits (Versant HCV RNA 3.0 b-DNA and Amplicor HCV Monitor), that also employ the WHO standard, allowed validation of the TaqMan assay against all major HCV genotypes. Both commercial methods detected HCV RNA over a wide dynamic range, but showed a consistent difference of about 0.3 log10 when evaluating samples of different HCV genotypes. The genome titers obtained with the three methods correlated with the infectivity titers previously determined for the HCV reference strains. TaqMan assays have become an essential tool to follow viral load in clinical samples and cell culture-based experiments and this technology offers significant advantages in linear dynamic range, sensitivity and customization.
(c) 2007 Wiley-Liss, Inc.
Figures

References
-
- Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. - PubMed
-
- Anderson JC, Simonetti J, Fisher DG, Williams J, Yamamura Y, Rodriguez N, Sullivan DG, Gretch DR, McMahon B, Williams KJ. Comparison of different HCV viral load and genotyping assays. J Clin Virol. 2003;28:27–37. - PubMed
-
- Bresters D, Cuypers HT, Reesink HW, Mauser-Bunschoten EP, van den Berg HM, Schaasberg WP, Wilber JC, Urdea MS, Neuwald P, Lelie PN. Comparison of quantitative cDNA-PCR with the branched DNA hybridization assay for monitoring plasma hepatitis C virus RNA levels in haemophilia patients participating in a controlled interferon trial. J Med Virol. 1994;43:262–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

