Using protein complexes to predict phenotypic effects of gene mutation
- PMID: 18042286
- PMCID: PMC2258176
- DOI: 10.1186/gb-2007-8-11-r252
Using protein complexes to predict phenotypic effects of gene mutation
Abstract
Background: Predicting the phenotypic effects of mutations is a central goal of genetics research; it has important applications in elucidating how genotype determines phenotype and in identifying human disease genes.
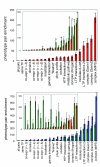
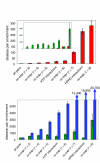
Results: Using a wide range of functional genomic data from the yeast Saccharomyces cerevisiae, we show that the best predictor of a protein's knockout phenotype is the knockout phenotype of other proteins that are present in a protein complex with it. Even the addition of multiple datasets does not improve upon the predictions made from protein complex membership. Similarly, we find that a proxy for protein complexes is a powerful predictor of disease phenotypes in humans.
Conclusion: We propose that identifying human protein complexes containing known disease genes will be an efficient method for large-scale disease gene discovery, and that yeast may prove to be an informative model system for investigating, and even predicting, the genetic basis of both Mendelian and complex disease phenotypes.
Figures
References
-
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, et al. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

