Characterization of Smoc-1 uncovers two transcript variants showing differential tissue and age specific expression in Bubalus bubalis
- PMID: 18042303
- PMCID: PMC2235864
- DOI: 10.1186/1471-2164-8-436
Characterization of Smoc-1 uncovers two transcript variants showing differential tissue and age specific expression in Bubalus bubalis
Abstract
Background: Secreted modular calcium binding protein-1 (Smoc-1) belongs to the BM-40 family which has been implicated with tissue remodeling, angiogenesis and bone mineralization. Besides its anticipated role in embryogenesis, Smoc-1 has been characterized only in a few mammalian species. We made use of the consensus sequence (5' CACCTCTCCACCTGCC 3') of 33.15 repeat loci to explore the buffalo transcriptome and uncovered the Smoc-1 transcript tagged with this repeat. The main objective of this study was to gain an insight into its structural and functional organization, and expressional status of Smoc-1 in water buffalo, Bubalus bubalis.
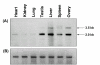
Results: We cloned and characterized the buffalo Smoc-1, including its copy number status, in-vitro protein expression, tissue & age specific transcription/translation, chromosomal mapping and localization to the basement membrane zone. Buffalo Smoc-1 was found to encode a secreted matricellular glycoprotein containing two EF-hand calcium binding motifs homologous to that of BM-40/SPARC family. In buffalo, this single copy gene consisted of 12 exons and was mapped onto the acrocentric chromosome 11. Though this gene was found to be evolutionarily conserved, the buffalo Smoc-1 showed conspicuous nucleotide/amino acid changes altering its secondary structure compared to that in other mammals. In silico analysis of the Smoc-1 proposed its glycoprotein nature with a calcium dependent conformation. Further, we unveiled two transcript variants of this gene, varying in their 3'UTR lengths but both coding for identical protein(s). Smoc-1 evinced highest expression of both the variants in liver and modest to negligible in other tissues. The relative expression of variant-02 was markedly higher compared to that of variant-01 in all the tissues examined. Moreover, expression of Smoc-1, though modest during the early ages, was conspicuously enhanced after 1 year and remained consistently higher during the entire life span of buffalo with gradual increment in expression of variant-02. Immunohistochemically, Smoc-1 was localized in the basement membrane zones and extracellular matrices of various tissues.
Conclusion: These data added to our understandings about the tissue, age and species specific functions of the Smoc-1. It also enabled us to demonstrate varying expression of the two transcript variants of Smoc-1 amongst different somatic tissues/gonads and ages, in spite of their identical coding frames. Pursuance of these variants for their roles in various disease phenotypes such as hepatocellular carcinoma and angiogenesis is envisaged to establish broader biological significance of this gene.
Figures





References
-
- Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8:163–173. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

