Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex
- PMID: 18042458
- PMCID: PMC2194599
- DOI: 10.1016/j.molcel.2007.11.001
Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex
Abstract
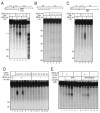
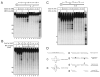
Mre11/Rad50 complexes in all organisms function in the repair of DNA double-strand breaks. In budding yeast, genetic evidence suggests that the Sae2 protein is essential for the processing of hairpin DNA intermediates and meiotic double-strand breaks by Mre11/Rad50 complexes, but the biochemical basis of this functional relationship is not known. Here we demonstrate that recombinant Sae2 binds DNA and exhibits endonuclease activity on single-stranded DNA independently of Mre11/Rad50 complexes, but hairpin DNA structures are cleaved cooperatively in the presence of Mre11/Rad50 or Mre11/Rad50/Xrs2. Hairpin structures are not processed at the tip by Sae2 but rather at single-stranded DNA regions adjacent to the hairpin. Truncation and missense mutants of Sae2 inactivate this endonuclease activity in vitro and fail to complement Deltasae2 strains in vivo for meiosis and recombination involving hairpin intermediates, suggesting that the catalytic activities of Sae2 are important for its biological functions.
Figures




References
-
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

