Characterization of an exosite binding inhibitor of matrix metalloproteinase 13
- PMID: 18042679
- PMCID: PMC2144595
- DOI: 10.1110/ps.073130208
Characterization of an exosite binding inhibitor of matrix metalloproteinase 13
Abstract
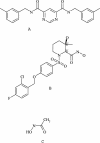
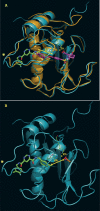
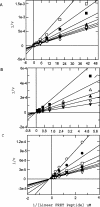
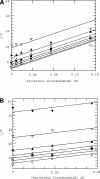
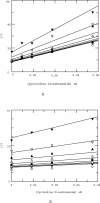
Matrix metalloproteinase 13 (MMP13) is a key enzyme implicated in the degradation of the extracellular matrix in osteoarthritis. Clinical administration of broad spectrum MMP inhibitors such as marimastat has been implicated in severe musculo-skeletal side effects. Consequently, research has been focused on designing inhibitors that selectively inhibit MMP13, thereby circumventing musculo-skeletal toxicities. A series of pyrimidine dicarboxamides were recently shown to be highly selective inhibitors of MMP13 with a novel binding mode. We have applied a molecular ruler to this exosite by dual inhibition studies involving a potent dicarboxamide in the presence of two metal chelators of different sizes. A larger hydroxamate mimic overlaps and antagonizes binding of the dicarboxamide to the exosite whereas the much smaller acetohydroxamate synergizes with the dicarboxamide. These studies elucidate the steric requirement for compounds that fit exclusively into the active site, a mandate for generating highly selective MMP13 inhibitors.
Figures
References
-
- Bigg, H.F., Rowan, A.D. The inhibition of metalloproteinases as a therapeutic target in rheumatoid arthritis and osteoarthritis. Curr. Opin. Pharmacol. 2001;1:314–320. - PubMed
-
- Brown, P.D. Ongoing trials with matrix metalloproteinase inhibitors. Expert Opin. Investig. Drugs. 2000;9:2167–2177. - PubMed
-
- Engel, C.K., Pirard, B., Schimanski, S., Kirsch, R., Habermann, J., Klinger, O., Schlotte, V., Weithmann, K.U., Wendt, K.U. Structural basis for the highly selective inhibition of MMP13. Chem. Biol. 2005;12:181–189. - PubMed
-
- Freskos, J.N., McDonald, J.J., Mischke, B.V., Mullins, P.B., Shieh, H.S., Stegeman, R.A., Stevens, A.M. Synthesis and identification of conformationally constrained selective MMP inhibitors. Bioorg. Med. Chem. Lett. 1999;9:1757–1760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
