A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide
- PMID: 18042713
- PMCID: PMC2154452
- DOI: 10.1073/pnas.0708102104
A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide
Abstract
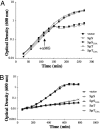
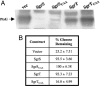
SgrS is a 227-nt small RNA that is expressed in Escherichia coli during glucose-phosphate stress, a condition associated with intracellular accumulation of glucose-6-phosphate caused by disruption of glycolytic flux. Under stress conditions, SgrS negatively regulates translation and stability of the ptsG mRNA, encoding the major glucose transporter, by means of a base pairing-dependent mechanism requiring the RNA chaperone Hfq. SgrS activity mitigates the effects of glucose-phosphate stress, and the present study has elucidated a function of SgrS that is proposed to contribute to the stress response. The 5' end of SgrS, upstream of the nucleotides involved in base pairing with the ptsG mRNA, contains a 43-aa ORF, sgrT, that is conserved in most species that contain SgrS-like small RNAs. The sgrT gene is translated in E. coli under conditions of glucose-phosphate stress. Analysis of alleles that separate the base pairing function of SgrS from the sgrT coding sequence revealed that either of these functions alone are sufficient for previously characterized SgrS phenotypes. SgrS-dependent down-regulation of ptsG mRNA stability does not require SgrT and SgrT by itself has no effect on ptsG mRNA stability. Cells expressing sgrT alone had a defect in glucose uptake even though they had nearly wild-type levels of PtsG (IICB(Glc)). Together, these data suggest that SgrS represents a previously unrecognized paradigm for small RNA (sRNA) regulators as a bifunctional RNA that encodes physiologically redundant but mechanistically distinct functions contributing to the same stress response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



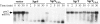
Comment in
-
Small RNAs making a small protein.Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20149-50. doi: 10.1073/pnas.0710634105. Epub 2007 Dec 11. Proc Natl Acad Sci U S A. 2007. PMID: 18077354 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

