Sequence-based analysis of pQBR103; a representative of a unique, transfer-proficient mega plasmid resident in the microbial community of sugar beet
- PMID: 18043644
- PMCID: PMC2656933
- DOI: 10.1038/ismej.2007.47
Sequence-based analysis of pQBR103; a representative of a unique, transfer-proficient mega plasmid resident in the microbial community of sugar beet
Abstract
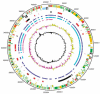
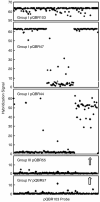
The plasmid pQBR103 was found within Pseudomonas populations colonizing the leaf and root surfaces of sugar beet plants growing at Wytham, Oxfordshire, UK. At 425 kb it is the largest self-transmissible plasmid yet sequenced from the phytosphere. It is known to enhance the competitive fitness of its host, and parts of the plasmid are known to be actively transcribed in the plant environment. Analysis of the complete sequence of this plasmid predicts a coding sequence (CDS)-rich genome containing 478 CDSs and an exceptional degree of genetic novelty; 80% of predicted coding sequences cannot be ascribed a function and 60% are orphans. Of those to which function could be assigned, 40% bore greatest similarity to sequences from Pseudomonas spp, and the majority of the remainder showed similarity to other gamma-proteobacterial genera and plasmids. pQBR103 has identifiable regions presumed responsible for replication and partitioning, but despite being tra+ lacks the full complement of any previously described conjugal transfer functions. The DNA sequence provided few insights into the functional significance of plant-induced transcriptional regions, but suggests that 14% of CDSs may be expressed (11 CDSs with functional annotation and 54 without), further highlighting the ecological importance of these novel CDSs. Comparative analysis indicates that pQBR103 shares significant regions of sequence with other plasmids isolated from sugar beet plants grown at the same geographic location. These plasmid sequences indicate there is more novelty in the mobile DNA pool accessible to phytosphere pseudomonas than is currently appreciated or understood.
Figures


Similar articles
-
The indigenous Pseudomonas plasmid pQBR103 encodes plant-inducible genes, including three putative helicases.FEMS Microbiol Ecol. 2004 Dec 27;51(1):9-17. doi: 10.1016/j.femsec.2004.07.006. FEMS Microbiol Ecol. 2004. PMID: 16329852
-
Impact of Plasmid pQBR103 Acquisition and Carriage on the Phytosphere Fitness of Pseudomonas fluorescens SBW25: Burden and Benefit.Appl Environ Microbiol. 1997 Apr;63(4):1584-7. doi: 10.1128/aem.63.4.1584-1587.1997. Appl Environ Microbiol. 1997. PMID: 16535581 Free PMC article.
-
Genetic characterization of Pseudomonas fluorescens SBW25 rsp gene expression in the phytosphere and in vitro.J Bacteriol. 2005 Dec;187(24):8477-88. doi: 10.1128/JB.187.24.8477-8488.2005. J Bacteriol. 2005. PMID: 16321952 Free PMC article.
-
Functional and phylogenetic analysis of a plant-inducible oligoribonuclease (orn) gene from an indigenous Pseudomonas plasmid.Microbiology (Reading). 2004 Sep;150(Pt 9):2889-2898. doi: 10.1099/mic.0.27250-0. Microbiology (Reading). 2004. PMID: 15347748
-
The evolution of IncP catabolic plasmids.Curr Opin Biotechnol. 2005 Jun;16(3):291-8. doi: 10.1016/j.copbio.2005.04.002. Curr Opin Biotechnol. 2005. PMID: 15961030 Review.
Cited by
-
Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96.Antimicrob Agents Chemother. 2013 Aug;57(8):3775-82. doi: 10.1128/AAC.00423-13. Epub 2013 May 28. Antimicrob Agents Chemother. 2013. PMID: 23716048 Free PMC article.
-
Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects.Environ Microbiol. 2015 Dec;17(12):5008-22. doi: 10.1111/1462-2920.12901. Epub 2015 Jun 25. Environ Microbiol. 2015. PMID: 25969927 Free PMC article.
-
Pmr, a histone-like protein H1 (H-NS) family protein encoded by the IncP-7 plasmid pCAR1, is a key global regulator that alters host function.J Bacteriol. 2010 Sep;192(18):4720-31. doi: 10.1128/JB.00591-10. Epub 2010 Jul 16. J Bacteriol. 2010. PMID: 20639326 Free PMC article.
-
A Post-segregational Killing Mechanism for Maintaining Plasmid PMF1 in Its Myxococcus fulvus Host.Front Cell Infect Microbiol. 2018 Aug 7;8:274. doi: 10.3389/fcimb.2018.00274. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30131946 Free PMC article.
-
Bacteriophages limit the existence conditions for conjugative plasmids.mBio. 2015 Jun 2;6(3):e00586. doi: 10.1128/mBio.00586-15. mBio. 2015. PMID: 26037122 Free PMC article.
References
-
- Bailey MJ, Kobayashi N, Lilley AK, Powell BJ, Thompson IP. Potential for gene transfer in the phytosphere: isolation and characterisation of naturally occurring plasmids. In: Bazin MJ, Lynch JM, editors. Environmental Gene Release. Chapman & Hall; London, UK: 1994. pp. 77–98.
-
- Bailey MJ, Lilley AK, Diaper JD. Gene transfer in the phyllosphere. In: Morris CE, Nicot P, Nguyen-the C, editors. Microbiology of Aerial Plant Surfaces. Plenum Publishing; New York, USA: 1996. pp. 103–123.
-
- Bailey MJ, Rainey PB, Zhang X-X, Lilley AK. Population dynamics, gene transfer and gene expression in plasmids, the role of the horizontal gene pool in local adaptation at the plant surface. In: Lindow SE, Hecht-Poinar I, Elliott VJ, editors. Microbiology of Aerial Plant Surfaces. American Phytopath. Soc. Press; St Paul, USA: 2001. pp. 171–189.
-
- Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2003;27:355–384. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

