Physical analyses of E. coli heteroduplex recombination products in vivo: on the prevalence of 5' and 3' patches
- PMID: 18043749
- PMCID: PMC2082072
- DOI: 10.1371/journal.pone.0001242
Physical analyses of E. coli heteroduplex recombination products in vivo: on the prevalence of 5' and 3' patches
Abstract
Background: Homologous recombination in Escherichia coli creates patches (non-crossovers) or splices (half crossovers), each of which may have associated heteroduplex DNA. Heteroduplex patches have recombinant DNA in one strand of the duplex, with parental flanking markers. Which DNA strand is exchanged in heteroduplex patches reflects the molecular mechanism of recombination. Several models for the mechanism of E. coli RecBCD-mediated recombinational double-strand-end (DSE) repair specify that only the 3'-ending strand invades the homologous DNA, forming heteroduplex in that strand. There is, however, in vivo evidence that patches are found in both strands.
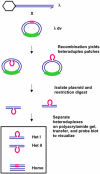
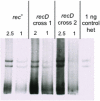
Methodology/principle findings: This paper re-examines heteroduplex-patch-strand polarity using phage lambda and the lambdadv plasmid as DNA substrates recombined via the E. coli RecBCD system in vivo. These DNAs are mutant for lambda recombination functions, including orf and rap, which were functional in previous studies. Heteroduplexes are isolated, separated on polyacrylamide gels, and quantified using Southern blots for heteroduplex analysis. This method reveals that heteroduplexes are still found in either 5' or 3' DNA strands in approximately equal amounts, even in the absence of orf and rap. Also observed is an independence of the RuvC Holliday-junction endonuclease on patch formation, and a slight but statistically significant alteration of patch polarity by recD mutation.
Conclusions/significance: These results indicate that orf and rap did not contribute to the presence of patches, and imply that patches occurring in both DNA strands reflects the molecular mechanism of recombination in E. coli. Most importantly, the lack of a requirement for RuvC implies that endonucleolytic resolution of Holliday junctions is not necessary for heteroduplex-patch formation, contrary to predictions of all of the major previous models. This implies that patches are not an alternative resolution of the same intermediate that produces splices, and do not bear on models for splice formation. We consider two mechanisms that use DNA replication instead of endonucleolytic resolution for formation of heteroduplex patches in either DNA strand: synthesis-dependent-strand annealing and a strand-assimilation mechanism.
Conflict of interest statement
Figures







Similar articles
-
Primary products of break-induced recombination by Escherichia coli RecE pathway.J Bacteriol. 1995 Apr;177(7):1692-8. doi: 10.1128/jb.177.7.1692-1698.1995. J Bacteriol. 1995. PMID: 7896689 Free PMC article.
-
Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3'-ending strand.Proc Natl Acad Sci U S A. 1998 Jun 9;95(12):6909-14. doi: 10.1073/pnas.95.12.6909. Proc Natl Acad Sci U S A. 1998. PMID: 9618512 Free PMC article.
-
Heteroduplex strand-specificity in restriction-stimulated recombination by the RecE pathway of Escherichia coli.Genetics. 1993 Mar;133(3):439-48. doi: 10.1093/genetics/133.3.439. Genetics. 1993. PMID: 8384141 Free PMC article.
-
Biological roles of the Escherichia coli RuvA, RuvB and RuvC proteins revealed.Mol Microbiol. 1992 Oct;6(19):2755-9. doi: 10.1111/j.1365-2958.1992.tb01454.x. Mol Microbiol. 1992. PMID: 1435254 Review.
-
Molecular mechanisms of Holliday junction processing in Escherichia coli.Adv Biophys. 1995;31:49-65. doi: 10.1016/0065-227x(95)99382-y. Adv Biophys. 1995. PMID: 7625278 Review.
Cited by
-
Modelling of crowded polymers elucidate effects of double-strand breaks in topological domains of bacterial chromosomes.Nucleic Acids Res. 2013 Aug;41(14):6808-15. doi: 10.1093/nar/gkt480. Epub 2013 Jun 5. Nucleic Acids Res. 2013. PMID: 23742906 Free PMC article.
-
RecBCD enzyme: mechanistic insights from mutants of a complex helicase-nuclease.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004123. doi: 10.1128/mmbr.00041-23. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047637 Free PMC article. Review.
-
Mu insertions are repaired by the double-strand break repair pathway of Escherichia coli.PLoS Genet. 2012;8(4):e1002642. doi: 10.1371/journal.pgen.1002642. Epub 2012 Apr 12. PLoS Genet. 2012. PMID: 22511883 Free PMC article.
References
-
- Myers RS, Stahl FW. χ and RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. - PubMed
-
- Palas KM, Kushner SR. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J Biol Chem. 1990;265:3447–3454. - PubMed
-
- Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. - PubMed
-
- Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of the Escherichia coli RecBCD enzyme. J Mol Biol. 1993;229:67–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

