An intronic signal for alternative splicing in the human genome
- PMID: 18043753
- PMCID: PMC2082412
- DOI: 10.1371/journal.pone.0001246
An intronic signal for alternative splicing in the human genome
Abstract
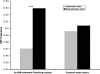
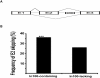
An important level at which the expression of programmed cell death (PCD) genes is regulated is alternative splicing. Our previous work identified an intronic splicing regulatory element in caspase-2 (casp-2) gene. This 100-nucleotide intronic element, In100, consists of an upstream region containing a decoy 3' splice site and a downstream region containing binding sites for splicing repressor PTB. Based on the signal of In100 element in casp-2, we have detected the In100-like sequences as a family of sequence elements associated with alternative splicing in the human genome by using computational and experimental approaches. A survey of human genome reveals the presence of more than four thousand In100-like elements in 2757 genes. These In100-like elements tend to locate more frequent in intronic regions than exonic regions. EST analyses indicate that the presence of In100-like elements correlates with the skipping of their immediate upstream exons, with 526 genes showing exon skipping in such a manner. In addition, In100-like elements are found in several human caspase genes near exons encoding the caspase active domain. RT-PCR experiments show that these caspase genes indeed undergo alternative splicing in a pattern predicted to affect their functional activity. Together, these results suggest that the In100-like elements represent a family of intronic signals for alternative splicing in the human genome.
Conflict of interest statement
Figures




References
-
- Johnson JM, Castle J, Garrett-Engele P, Kan ZY, Loerch PM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. - PubMed
-
- Wu JY, Tang H, Havlioglu N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog Mol Subcell Biol. 2003;31:153–85. - PubMed
-
- Wu JY, Yuan L, Havlioglu N. Alternatively spliced genes. In Encyclopedia of Molecular Cell Biology and Molecular Medicine Vol1. pp 125–177. Edited by Meyers R.A. (2nd edition) (Wiley-VCH) 2004
-
- Boyce M, Degterev A, Yuan J. Caspases: an ancient cellular sword of Damocles. Cell Death Differ. 2004;11:29–37. - PubMed
-
- Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

