Post-transcriptional gene regulation: from genome-wide studies to principles
- PMID: 18043867
- PMCID: PMC2771128
- DOI: 10.1007/s00018-007-7447-6
Post-transcriptional gene regulation: from genome-wide studies to principles
Abstract
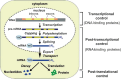
Post-transcriptional regulation of gene expression plays important roles in diverse cellular processes such as development, metabolism and cancer progression. Whereas many classical studies explored the mechanistics and physiological impact on specific mRNA substrates, the recent development of genome-wide analysis tools enables the study of post-transcriptional gene regulation on a global scale. Importantly, these studies revealed distinct programs of RNA regulation, suggesting a complex and versatile post-transcriptional regulatory network. This network is controlled by specific RNA-binding proteins and/or non-coding RNAs, which bind to specific sequence or structural elements in the RNAs and thereby regulate subsets of mRNAs that partly encode functionally related proteins. It will be a future challenge to link the spectra of targets for RNA-binding proteins to post-transcriptional regulatory programs and to reveal its physiological implications.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

