Phylogenomics and signature proteins for the alpha proteobacteria and its main groups
- PMID: 18045498
- PMCID: PMC2241609
- DOI: 10.1186/1471-2180-7-106
Phylogenomics and signature proteins for the alpha proteobacteria and its main groups
Abstract
Background: Alpha proteobacteria are one of the largest and most extensively studied groups within bacteria. However, for these bacteria as a whole and for all of its major subgroups (viz. Rhizobiales, Rhodobacterales, Rhodospirillales, Rickettsiales, Sphingomonadales and Caulobacterales), very few or no distinctive molecular or biochemical characteristics are known.
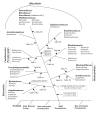
Results: We have carried out comprehensive phylogenomic analyses by means of Blastp and PSI-Blast searches on the open reading frames in the genomes of several alpha-proteobacteria (viz. Bradyrhizobium japonicum, Brucella suis, Caulobacter crescentus, Gluconobacter oxydans, Mesorhizobium loti, Nitrobacter winogradskyi, Novosphingobium aromaticivorans, Rhodobacter sphaeroides 2.4.1, Silicibacter sp. TM1040, Rhodospirillum rubrum and Wolbachia (Drosophila) endosymbiont). These studies have identified several proteins that are distinctive characteristics of all alpha-proteobacteria, as well as numerous proteins that are unique repertoires of all of its main orders (viz. Rhizobiales, Rhodobacterales, Rhodospirillales, Rickettsiales, Sphingomonadales and Caulobacterales) and many families (viz. Rickettsiaceae, Anaplasmataceae, Rhodospirillaceae, Acetobacteraceae, Bradyrhiozobiaceae, Brucellaceae and Bartonellaceae). Many other proteins that are present at different phylogenetic depths in alpha-proteobacteria provide important information regarding their evolution. The evolutionary relationships among alpha-proteobacteria as deduced from these studies are in excellent agreement with their branching pattern in the phylogenetic trees and character compatibility cliques based on concatenated sequences for many conserved proteins. These studies provide evidence that the major groups within alpha-proteobacteria have diverged in the following order: (Rickettsiales(Rhodospirillales (Sphingomonadales (Rhodobacterales (Caulobacterales-Parvularculales (Rhizobiales)))))). We also describe two conserved inserts in DNA Gyrase B and RNA polymerase beta subunit that are distinctive characteristics of the Sphingomonadales and Rhodosprilllales species, respectively. The results presented here also provide support for the grouping of Hyphomonadaceae and Parvularcula species with the Caulobacterales and the placement of Stappia aggregata with the Rhizobiaceae group.
Conclusion: The alpha-proteobacteria-specific proteins and indels described here provide novel and powerful means for the taxonomic, biochemical and molecular biological studies on these bacteria. Their functional studies should prove helpful in identifying novel biochemical and physiological characteristics that are unique to these bacteria.
Figures




Similar articles
-
Protein signatures distinctive of alpha proteobacteria and its subgroups and a model for alpha-proteobacterial evolution.Crit Rev Microbiol. 2005;31(2):101-35. doi: 10.1080/10408410590922393. Crit Rev Microbiol. 2005. PMID: 15986834 Review.
-
Signature proteins that are distinctive of alpha proteobacteria.BMC Genomics. 2005 Jun 16;6:94. doi: 10.1186/1471-2164-6-94. BMC Genomics. 2005. PMID: 15960851 Free PMC article.
-
A robust species tree for the alphaproteobacteria.J Bacteriol. 2007 Jul;189(13):4578-86. doi: 10.1128/JB.00269-07. Epub 2007 May 4. J Bacteriol. 2007. PMID: 17483224 Free PMC article.
-
Application of the character compatibility approach to generalized molecular sequence data: branching order of the proteobacterial subdivisions.J Mol Evol. 2007 Jan;64(1):90-100. doi: 10.1007/s00239-006-0082-2. Epub 2006 Dec 9. J Mol Evol. 2007. PMID: 17160641
-
Molecular signatures (unique proteins and conserved indels) that are specific for the epsilon proteobacteria (Campylobacterales).BMC Genomics. 2006 Jul 4;7:167. doi: 10.1186/1471-2164-7-167. BMC Genomics. 2006. PMID: 16817973 Free PMC article. Review.
Cited by
-
CceR and AkgR regulate central carbon and energy metabolism in alphaproteobacteria.mBio. 2015 Feb 3;6(1):e02461-14. doi: 10.1128/mBio.02461-14. mBio. 2015. PMID: 25650399 Free PMC article.
-
Comparative genomics analysis reveals genetic characteristics and nitrogen fixation profile of Bradyrhizobium.iScience. 2024 Jan 18;27(2):108948. doi: 10.1016/j.isci.2024.108948. eCollection 2024 Feb 16. iScience. 2024. PMID: 38322985 Free PMC article.
-
Signature proteins for the major clades of Cyanobacteria.BMC Evol Biol. 2010 Jan 25;10:24. doi: 10.1186/1471-2148-10-24. BMC Evol Biol. 2010. PMID: 20100331 Free PMC article.
-
Role of Caulobacter Cell Surface Structures in Colonization of the Air-Liquid Interface.J Bacteriol. 2019 Aug 22;201(18):e00064-19. doi: 10.1128/JB.00064-19. Print 2019 Sep 15. J Bacteriol. 2019. PMID: 31010900 Free PMC article.
-
Next-generation pyrosequencing analysis of microbial biofilm communities on granular activated carbon in treatment of oil sands process-affected water.Appl Environ Microbiol. 2015 Jun 15;81(12):4037-48. doi: 10.1128/AEM.04258-14. Epub 2015 Apr 3. Appl Environ Microbiol. 2015. PMID: 25841014 Free PMC article.
References
-
- Kersters K, Devos P, Gillis M, Swings J, Vandamme P, Stackebrandt E. Introduction to the Proteobacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH and Stackebrandt E, editor. The Prokaryotes: A Handbook on the Biology of Bacteria. 3rd edition, Release 3.12 . New York, Springer; 2006. pp. 3–37.
-
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappe MS, Short JM, Carrington JC, Mathur EJ. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. - PubMed
-
- Fredricks DN. Introduction to the Rickettsiales and other intracellular prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH and Stackebrandt E, editor. The Prokaryotes: A Handbook on the Biology of Bacteria. 3rd edition. New York, Springer; 2006. pp. 457–466.
-
- Skorpil P, Broughton WJ. Molecular interactions between Rhizobium and legumes. Prog Mol Subcell Biol. 2006;41:143–164. - PubMed
-
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

