The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously
- PMID: 18045837
- PMCID: PMC2277323
- DOI: 10.1242/dev.011288
The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously
Abstract
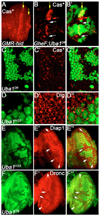
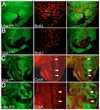
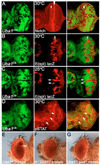
Ubiquitination is an essential process regulating turnover of proteins for basic cellular processes such as the cell cycle and cell death (apoptosis). Ubiquitination is initiated by ubiquitin-activating enzymes (E1), which activate and transfer ubiquitin to ubiquitin-conjugating enzymes (E2). Conjugation of target proteins with ubiquitin is then mediated by ubiquitin ligases (E3). Ubiquitination has been well characterized using mammalian cell lines and yeast genetics. However, the consequences of partial or complete loss of ubiquitin conjugation in a multi-cellular organism are not well understood. Here, we report the characterization of Uba1, the only E1 in Drosophila. We found that weak and strong Uba1 alleles behave genetically differently with sometimes opposing phenotypes. Whereas weak Uba1 alleles protect cells from cell death, clones of strong Uba1 alleles are highly apoptotic. Strong Uba1 alleles cause cell cycle arrest which correlates with failure to reduce cyclin levels. Surprisingly, clones of strong Uba1 mutants stimulate neighboring wild-type tissue to undergo cell division in a non-autonomous manner giving rise to overgrowth phenotypes of the mosaic fly. We demonstrate that the non-autonomous overgrowth is caused by failure to downregulate Notch signaling in Uba1 mutant clones. In summary, the phenotypic analysis of Uba1 demonstrates that impaired ubiquitin conjugation has significant consequences for the organism, and may implicate Uba1 as a tumor suppressor gene.
Figures




References
-
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. - PubMed
-
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev. Biol. 1989;136:346–362. - PubMed
-
- Cashio P, Lee TV, Bergmann A. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 2005;16:225–235. - PubMed
-
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 2003;10:892–898. - PubMed
-
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

