A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo
- PMID: 18045839
- PMCID: PMC2205987
- DOI: 10.1242/dev.010744
A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo
Abstract
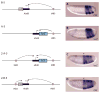
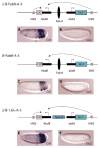
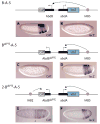
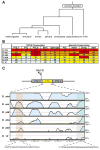
A key question in our understanding of the cis-regulation of gene expression during embryonic development has been the molecular mechanism that directs enhancers to specific promoters within a gene complex. Promoter competition and insulators are thought to play a role in regulating these interactions. In the bithorax complex of Drosophila, the IAB5 enhancer is located 55 kb 3' of the Abdominal-B (Abd-B) promoter and 48 kb 5' of the abdominal-A (abd-A) promoter. Although roughly equidistant from the two promoters, IAB5 specifically interacts only with the Abdominal-B promoter, even though the enhancer and promoter are separated by at least two insulators. Here we demonstrate that a 255 bp element, located 40 bp 5' of the Abd-B transcriptional start site, has a novel cis-regulatory activity as it is able to tether IAB5 to the Abd-B promoter in transgenic embryos. The tethering element is sufficient to direct IAB5 to an ectopic promoter in competition assays. Deletion of the promoter-tethering element results in the redirection of enhancer-driven gene expression on transgenes. Taken together, these results provide evidence that specific long-range enhancer-promoter interactions in the bithorax complex are regulated by a tethering element 5' of the Abd-B promoter. We discuss a bioinformatic analysis of the tethering element across different Drosophila species and a possible molecular mechanism by which this element functions. We also examine existing evidence that this novel class of cis-regulatory elements might regulate enhancer-promoter specificity at other gene complexes.
Figures



References
-
- Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev Biol. 2006;293:294–304. - PubMed
-
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

