RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe
- PMID: 18045993
- PMCID: PMC2230603
- DOI: 10.1091/mbc.e07-06-0618
RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe
Abstract
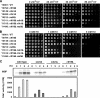
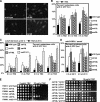
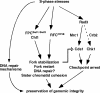
Sister chromatid cohesion is established during S phase near the replication fork. However, how DNA replication is coordinated with chromosomal cohesion pathway is largely unknown. Here, we report studies of fission yeast Ctf18, a subunit of the RFC(Ctf18) replication factor C complex, and Chl1, a putative DNA helicase. We show that RFC(Ctf18) is essential in the absence of the Swi1-Swi3 replication fork protection complex required for the S phase stress response. Loss of Ctf18 leads to an increased sensitivity to S phase stressing agents, a decreased level of Cds1 kinase activity, and accumulation of DNA damage during S phase. Ctf18 associates with chromatin during S phase, and it is required for the proper resumption of replication after fork arrest. We also show that chl1Delta is synthetically lethal with ctf18Delta and that a dosage increase of chl1(+) rescues sensitivities of swi1Delta to S phase stressing agents, indicating that Chl1 is involved in the S phase stress response. Finally, we demonstrate that inactivation of Ctf18, Chl1, or Swi1-Swi3 leads to defective centromere cohesion, suggesting the role of these proteins in chromosome segregation. We propose that RFC(Ctf18) and the Swi1-Swi3 complex function in separate and redundant pathways essential for replication fork stabilization to facilitate sister chromatid cohesion in fission yeast.
Figures





References
-
- Alfa C., Fantes P., Hyams J., McLeod M., Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993.
-
- Amann J., Valentine M., Kidd V. J., Lahti J. M. Localization of chi1-related helicase genes to human chromosome regions 12p11 and 12p 13, similarity between parts of these genes and conserved human telomeric-associated DNA. Genomics. 1996;32:260–265. - PubMed
-
- Aparicio O. M., Weinstein D. M., Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

