Application of phage display to high throughput antibody generation and characterization
- PMID: 18047641
- PMCID: PMC2258204
- DOI: 10.1186/gb-2007-8-11-r254
Application of phage display to high throughput antibody generation and characterization
Abstract
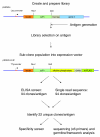
We have created a high quality phage display library containing over 1010 human antibodies and describe its use in the generation of antibodies on an unprecedented scale. We have selected, screened and sequenced over 38,000 recombinant antibodies to 292 antigens, yielding over 7,200 unique clones. 4,400 antibodies were characterized by specificity testing and detailed sequence analysis and the data/clones are available online. Sensitive detection was demonstrated in a bead based flow cytometry assay. Furthermore, positive staining by immunohistochemistry on tissue microarrays was found for 37% (143/381) of antibodies. Thus, we have demonstrated the potential of and illuminated the issues associated with genome-wide monoclonal antibody generation.
Figures





References
-
- Taussig MJ, Stoevesandt O, Borrebaeck CAK, Bradbury AR, Cahill D, Cambillau C, de Daruvar A, Dubel S, Eichler J, Frank R, et al. ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

