Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis
- PMID: 18052041
- PMCID: PMC2535579
- DOI: 10.1021/bi700970a
Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis
Abstract
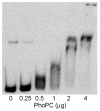
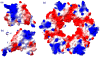
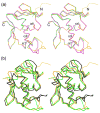
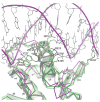
The PhoP-PhoR two-component signaling system from Mycobacterium tuberculosis is essential for the virulence of the tubercle bacillus. The response regulator, PhoP, regulates expression of over 110 genes. In order to elucidate the regulatory mechanism of PhoP, we determined the crystal structure of its DNA-binding domain (PhoPC). PhoPC exhibits a typical fold of the winged helix-turn-helix subfamily of response regulators. The structure starts with a four-stranded antiparallel beta-sheet, followed by a three-helical bundle of alpha-helices, and then a C-terminal beta-hairpin, which together with a short beta-strand between the first and second helices forms a three-stranded antiparallel beta-sheet. Structural elements are packed through a hydrophobic core, with the first helix providing a scaffold for the rest of the domain to pack. The second and third helices and the long, flexible loop between them form the helix-turn-helix motif, with the third helix being the recognition helix. The C-terminal beta-hairpin turn forms the wing motif. The molecular surfaces around the recognition helix and the wing residues show strong positive electrostatic potential, consistent with their roles in DNA binding and nucleotide sequence recognition. The crystal packing of PhoPC gives a hexamer ring, with neighboring molecules interacting in a head-to-tail fashion. This packing interface suggests that PhoPC could bind DNA in a tandem association. However, this mode of DNA binding is likely to be nonspecific because the recognition helix is partially blocked and would be prevented from inserting into the major groove of DNA. Detailed structural analysis and implications with respect to DNA binding are discussed.
Figures
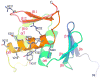

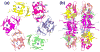
Similar articles
-
A single-amino-acid substitution in the C terminus of PhoP determines DNA-binding specificity of the virulence-associated response regulator from Mycobacterium tuberculosis.J Mol Biol. 2010 May 21;398(5):647-56. doi: 10.1016/j.jmb.2010.03.056. Epub 2010 Apr 2. J Mol Biol. 2010. PMID: 20363229
-
Domain structure of virulence-associated response regulator PhoP of Mycobacterium tuberculosis: role of the linker region in regulator-promoter interaction(s).J Biol Chem. 2010 Nov 5;285(45):34309-18. doi: 10.1074/jbc.M110.135822. Epub 2010 Sep 2. J Biol Chem. 2010. PMID: 20814030 Free PMC article.
-
Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.J Mol Biol. 2000 Feb 4;295(5):1225-36. doi: 10.1006/jmbi.1999.3379. J Mol Biol. 2000. PMID: 10653699
-
Structural diversity based on variability in quaternary association. A case study involving eubacterial and related SSBs.Methods Mol Biol. 2012;922:23-35. doi: 10.1007/978-1-62703-032-8_2. Methods Mol Biol. 2012. PMID: 22976175 Review.
-
RNA sculpting by the primordial Helix-clasp-Helix-Strand-Loop (HcH-SL) motif enforces chemical recognition enabling diverse KH domain functions.J Biol Chem. 2025 May;301(5):108474. doi: 10.1016/j.jbc.2025.108474. Epub 2025 Apr 2. J Biol Chem. 2025. PMID: 40185232 Free PMC article. Review.
Cited by
-
A Low-Prevalence Single-Nucleotide Polymorphism in the Sensor Kinase PhoR in Mycobacterium tuberculosis Suppresses Its Autophosphatase Activity and Reduces Pathogenic Fitness: Implications in Evolutionary Selection.Front Microbiol. 2021 Aug 25;12:724482. doi: 10.3389/fmicb.2021.724482. eCollection 2021. Front Microbiol. 2021. PMID: 34512602 Free PMC article.
-
Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis.Microbiol Mol Biol Rev. 2011 Dec;75(4):566-82. doi: 10.1128/MMBR.05004-11. Microbiol Mol Biol Rev. 2011. PMID: 22126994 Free PMC article. Review.
-
Withdrawn.Infect Disord Drug Targets. 2012 Nov 16. Online ahead of print. Infect Disord Drug Targets. 2012. PMID: 23167715 Free PMC article.
-
The atypical OmpR/PhoB response regulator ChxR from Chlamydia trachomatis forms homodimers in vivo and binds a direct repeat of nucleotide sequences.J Bacteriol. 2011 Jan;193(2):389-98. doi: 10.1128/JB.00833-10. Epub 2010 Nov 5. J Bacteriol. 2011. PMID: 21057008 Free PMC article.
-
Expression, purification, crystallization and preliminary X-ray analysis of the DNA-binding domain of a Chlamydia trachomatis OmpR/PhoB-subfamily response regulator homolog, ChxR.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Aug 1;65(Pt 8):791-4. doi: 10.1107/S1744309109025184. Epub 2009 Jul 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19652341 Free PMC article.
References
-
- Raviglione MC, Smith IM. XDR tuberculosis--implications for global public health. N Engl J Med. 2007;356:656–659. - PubMed
-
- Av-Gay Y, Deretic V. Two-component systems, protein kinases, and signal transduction in Mycobacterium tuberculosis. In: Cole ST, editor. Tuberculosis and the Tubercle Bacillus. ASM Press; Washington, D. C.: 2005. pp. 359–367.
-
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
-
- Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41:179–187. - PubMed
-
- Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60:312–330. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

