Avoidance of protein fold disruption in natural virus recombinants
- PMID: 18052529
- PMCID: PMC2092379
- DOI: 10.1371/journal.ppat.0030181
Avoidance of protein fold disruption in natural virus recombinants
Abstract
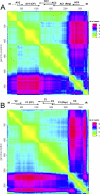
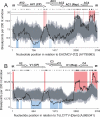
With the development of reliable recombination detection tools and an increasing number of available genome sequences, many studies have reported evidence of recombination in a wide range of virus genera. Recombination is apparently a major mechanism in virus evolution, allowing viruses to evolve more quickly by providing immediate direct access to many more areas of a sequence space than are accessible by mutation alone. Recombination has been widely described amongst the insect-transmitted plant viruses in the genus Begomovirus (family Geminiviridae), with potential recombination hot- and cold-spots also having been identified. Nevertheless, because very little is understood about either the biochemical predispositions of different genomic regions to recombine or what makes some recombinants more viable than others, the sources of the evolutionary and biochemical forces shaping distinctive recombination patterns observed in nature remain obscure. Here we present a detailed analysis of unique recombination events detectable in the DNA-A and DNA-A-like genome components of bipartite and monopartite begomoviruses. We demonstrate both that recombination breakpoint hot- and cold-spots are conserved between the two groups of viruses, and that patterns of sequence exchange amongst the genomes are obviously non-random. Using a computational technique designed to predict structural perturbations in chimaeric proteins, we demonstrate that observed recombination events tend to be less disruptive than sets of simulated ones. Purifying selection acting against natural recombinants expressing improperly folded chimaeric proteins is therefore a major determinant of natural recombination patterns in begomoviruses.
Conflict of interest statement
Figures
References
-
- Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. - PubMed
-
- Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature. 2006;443:89–92. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

