N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance
- PMID: 18052601
- PMCID: PMC2100143
- DOI: 10.1371/journal.pmed.0040335
N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance
Abstract
Background: The catalytically active 66-kDa subunit of the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) consists of DNA polymerase, connection, and ribonuclease H (RNase H) domains. Almost all known RT inhibitor resistance mutations identified to date map to the polymerase domain of the enzyme. However, the connection and RNase H domains are not routinely analysed in clinical samples and none of the genotyping assays available for patient management sequence the entire RT coding region. The British Columbia Centre for Excellence in HIV/AIDS (the Centre) genotypes clinical isolates up to codon 400 in RT, and our retrospective statistical analyses of the Centre's database have identified an N348I mutation in the RT connection domain in treatment-experienced individuals. The objective of this multidisciplinary study was to establish the in vivo relevance of this mutation and its role in drug resistance.
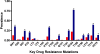
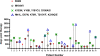
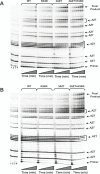
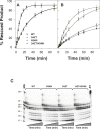
Methods and findings: The prevalence of N348I in clinical isolates, the time taken for it to emerge under selective drug pressure, and its association with changes in viral load, specific drug treatment, and known drug resistance mutations was analysed from genotypes, viral loads, and treatment histories from the Centre's database. N348I increased in prevalence from below 1% in 368 treatment-naïve individuals to 12.1% in 1,009 treatment-experienced patients (p = 7.7 x 10(-12)). N348I appeared early in therapy and was highly associated with thymidine analogue mutations (TAMs) M41L and T215Y/F (p < 0.001), the lamivudine resistance mutations M184V/I (p < 0.001), and non-nucleoside RTI (NNRTI) resistance mutations K103N and Y181C/I (p < 0.001). The association with TAMs and NNRTI resistance mutations was consistent with the selection of N348I in patients treated with regimens that included both zidovudine and nevirapine (odds ratio 2.62, 95% confidence interval 1.43-4.81). The appearance of N348I was associated with a significant increase in viral load (p < 0.001), which was as large as the viral load increases observed for any of the TAMs. However, this analysis did not account for the simultaneous selection of other RT or protease inhibitor resistance mutations on viral load. To delineate the role of this mutation in RT inhibitor resistance, N348I was introduced into HIV-1 molecular clones containing different genetic backbones. N348I decreased zidovudine susceptibility 2- to 4-fold in the context of wild-type HIV-1 or when combined with TAMs. N348I also decreased susceptibility to nevirapine (7.4-fold) and efavirenz (2.5-fold) and significantly potentiated resistance to these drugs when combined with K103N. Biochemical analyses of recombinant RT containing N348I provide supporting evidence for the role of this mutation in zidovudine and NNRTI resistance and give some insight into the molecular mechanism of resistance.
Conclusions: This study provides the first in vivo evidence that treatment with RT inhibitors can select a mutation (i.e., N348I) outside the polymerase domain of the HIV-1 RT that confers dual-class resistance. Its emergence, which can happen early during therapy, may significantly impact on a patient's response to antiretroviral therapies containing zidovudine and nevirapine. This study also provides compelling evidence for investigating the role of other mutations in the connection and RNase H domains in virological failure.
Conflict of interest statement
PRH has received honoraria, research and travel grants and served as a speaker from many pharmaceutical companies, and has served as a consultant to HIV drug resistance testing companies including Virco and Monogram Biosciences. SHY, CWS, JF, MZ, DT, VDL, BW, MK, NSC, and GT declare no competing interests.
Figures
Comment in
-
Should we include connection domain mutations of HIV-1 reverse transcriptase in HIV resistance testing.PLoS Med. 2007 Dec;4(12):e346. doi: 10.1371/journal.pmed.0040346. PLoS Med. 2007. PMID: 18052605 Free PMC article.
References
-
- Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. - PubMed
-
- Esnouf R, Ren J, Ross C, Jones Y, Stammers D, et al. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat Struct Biol. 1995;2:303–308. - PubMed
-
- Ding J, Das K, Moereels H, Koymans L, Andries K, et al. Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol. 1995;2:407–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

