An insight into the phylogenetic history of HOX linked gene families in vertebrates
- PMID: 18053128
- PMCID: PMC2235844
- DOI: 10.1186/1471-2148-7-239
An insight into the phylogenetic history of HOX linked gene families in vertebrates
Abstract
Background: The human chromosomes 2q, 7, 12q and 17q show extensive intra-genomic homology, containing duplicate, triplicate and quadruplicate paralogous regions centered on the HOX gene clusters. The fact that two or more representatives of different gene families are linked with HOX clusters is taken as evidence that these paralogous gene sets might have arisen from a single chromosomal segment through block or whole chromosome duplication events. This would imply that the constituent genes including the HOX clusters reflect the architecture of a single ancestral block (before vertebrate origin) where all of these genes were linked in a single copy.
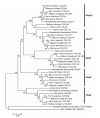
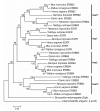
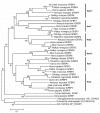
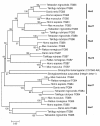
Results: In the present study we have employed the currently available set of protein data for a wide variety of vertebrate and invertebrate genomes to analyze the phylogenetic history of 11 multigene families with three or more of their representatives linked to human HOX clusters. A topology comparison approach revealed four discrete co-duplicated groups: group 1 involves the genes from GLI, HH, INHB, IGFBP (cluster-1), and SLC4A families; group 2 involves ERBB, ZNFN1A, and IGFBP (cluster-2) gene families; group 3 involves the HOX clusters and the SP gene family; group 4 involves the integrin beta chain and myosine light chain families. The distinct genes within each co-duplicated group share the same evolutionary history and are duplicated in concert with each other, while the constituent genes of two different co-duplicated groups may not share their evolutionary history and may not have duplicated simultaneously.
Conclusion: We conclude that co-duplicated groups may themselves be remnants of ancient small-scale duplications (involving chromosomal segments or gene-clusters) which occurred at different time points during chordate evolution. Whereas the recent combination of genes from distinct co-duplicated groups on different chromosomal regions (human chromosomes 2q, 7, 12q, and 17q) is probably the outcome of subsequent rearrangement of genomic segments, including syntenic groups of genes.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

