A new generation of pPRIG-based retroviral vectors
- PMID: 18053131
- PMCID: PMC2241607
- DOI: 10.1186/1472-6750-7-85
A new generation of pPRIG-based retroviral vectors
Abstract
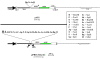
Background: Retroviral vectors are valuable tools for gene transfer. Particularly convenient are IRES-containing retroviral vectors expressing both the protein of interest and a marker protein from a single bicistronic mRNA. This coupled expression increases the relevance of tracking and/or selection of transduced cells based on the detection of a marker protein. pAP2 is a retroviral vector containing eGFP downstream of a modified IRES element of EMCV origin, and a CMV enhancer-promoter instead of the U3 region of the 5'LTR, which increases its efficiency in transient transfection. However, pAP2 contains a limited multicloning site (MCS) and shows weak eGFP expression, which previously led us to engineer an improved version, termed pPRIG, harboring: i) the wild-type ECMV IRES sequence, thereby restoring its full activity; ii) an optimized MCS flanked by T7 and SP6 sequences; and iii) a HA tag encoding sequence 5' of the MCS (pPRIG HAa/b/c).
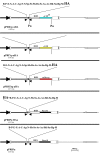
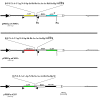
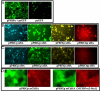
Results: The convenience of pPRIG makes it a good basic vector to generate additional derivatives for an extended range of use. Here we present several novel pPRIG-based vectors (collectively referred to as PRIGs) in which : i) the HA tag sequence was inserted in the three reading frames 3' of the MCS (3'HA PRIGs); ii) a functional domain (ER, VP16 or KRAB) was inserted either 5' or 3' of the MCS (<< modular >> PRIGs); iii) eGFP was replaced by either eCFP, eYFP, mCherry or puro-R (<< single color/resistance >> PRIGs); and iv) mCherry, eYFP or eGFP was inserted 5' of the MCS of the IRES-eGFP, IRES-eCFP or IRES-Puro-R containing PRIGs, respectively (<< dual color/selection >> PRIGs). Additionally, some of these PRIGs were also constructed in a pMigR MSCV background which has been widely used in pluripotent cells.
Conclusion: These novel vectors allow for straightforward detection of any expressed protein (3'HA PRIGs), for functional studies of chimeric proteins (<< modular >> PRIGs), for multiple transductions and fluorescence analyses of transduced cells (<< single color/resistance >> PRIGs), or for quantitative detection of studied proteins in independently identified/selected transduced cells (<< dual color/selection >> PRIGs). They maintain the original advantages of pPRIG and provide suitable tools for either transient or stable expression and functional studies in a large range of experimental settings.
Figures



References
-
- Galipeau J, Li H, Paquin A, Sicilia F, Karpati G, Nalbantoglu J. Vesicular stomatitis virus G pseudotyped retrovector mediates effective in vivo suicide gene delivery in experimental brain cancer. Cancer Res. 1999;59:2384–2394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

