Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development
- PMID: 18053265
- PMCID: PMC2217549
- DOI: 10.1186/1471-213X-7-135
Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development
Abstract
Background: Dopaminergic neurons form in diverse areas of the vertebrate di- and mesencephalon to constitute several major neuromodulatory systems. While much is known about mammalian mesencephalic dopaminergic neuron development, little is known about the specification of the diencephalic dopaminergic groups. The transcription factors Pitx3 and Lmx1b play an important role in mammalian mesencephalic dopaminergic specification, and Nurr1/Nr4a2 has been shown to contribute to specification of the dopaminergic neurotransmitter phenotype. We use zebrafish to analyze potentially evolutionarily conserved roles of these transcription factors in a vertebrate brain that lacks a mesencephalic dopaminergic system, but has an ascending dopaminergic system in the ventral diencephalon.
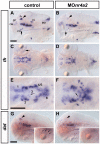
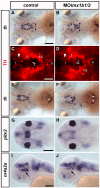
Results: We use a combination of fluorescent in situ hybridization and immunohistochemistry to determine whether nr4a2, lmx1b, and pitx3 genes are expressed in mature dopaminergic neurons or in potential precursor populations. We identify a second nr4a2 paralogue, nr4a2a, and find it co-expressed with Tyrosine hydroxylase in preoptic, pretectal and retinal amacrine dopaminergic neurons, while nr4a2b is only expressed in preoptic and retinal dopaminergic neurons. Both zebrafish nr4a2 paralogues are not expressed in ventral diencephalic dopaminergic neurons with ascending projections. Combined morpholino antisense oligo mediated knock-down of both nr4a2a and nr4a2b transcripts reveals that all zebrafish dopaminergic neurons expressing nr4a2a depend on Nr4a2 activity for tyrosine hydroxylase and dopamine transporter expression. Zebrafish lmx1b.1 is expressed in noradrenergic neurons of the locus coeruleus and medulla oblongata, but knock-down reveals that it is specifically required for tyrosine hydroxylase expression only in the medulla oblongata area postrema noradrenergic neurons. Both lmx1b genes and pitx3 are not expressed in dopaminergic neurons, but in a diencephalic territory that might contain precursor cells for ventral diencephalic dopaminergic neurons. Upon morpholino knock-down of both lmx1b paralogues, the number of neurons in diencephalic dopaminergic clusters with ascending projections appears specifically reduced. Thus lmx1b paralogues may contribute to the generation of diencephalic dopaminergic precursors. Conversely, knock-down of pitx3 does not specifically affect any diencephalic DA cluster.
Conclusion: Our data indicate a conserved evolutionary role of Nr4a2 proteins in specification of the neurotransmitter phenotype, albeit it appears to be only one of several regulatory modules of dopaminergic differentiation, as most ventral diencephalic dopaminergic neurons do not express nr4a2 genes in zebrafish. For zebrafish lmx1b genes, which are not expressed in mature dopaminergic neurons, our data suggest a role in diencephalic precursor populations contributing to the ascending dopaminergic systems. A di-mesencephalic longitudinal domain of lmx1b expression may be the basis for the expansion and posterior shift of ventral di-/mesencephalic dopaminergic populations with ascending projections during evolution.
Figures

Similar articles
-
In vivo expression of Nurr1/Nr4a2a in developing retinal amacrine subtypes in zebrafish Tg(nr4a2a:eGFP) transgenics.J Comp Neurol. 2017 Jun 1;525(8):1962-1979. doi: 10.1002/cne.24185. Epub 2017 Mar 15. J Comp Neurol. 2017. PMID: 28177524
-
Nr4a2 is essential for the differentiation of dopaminergic neurons during zebrafish embryogenesis.Mol Cell Neurosci. 2008 Oct;39(2):202-10. doi: 10.1016/j.mcn.2008.06.010. Epub 2008 Jun 25. Mol Cell Neurosci. 2008. PMID: 18638558
-
Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development.Curr Biol. 2007 May 15;17(10):873-80. doi: 10.1016/j.cub.2007.04.003. Epub 2007 May 3. Curr Biol. 2007. PMID: 17481897
-
The role of Pitx3 in survival of midbrain dopaminergic neurons.J Neural Transm Suppl. 2006;(70):57-60. doi: 10.1007/978-3-211-45295-0_10. J Neural Transm Suppl. 2006. PMID: 17017509 Review.
-
Terminal differentiation ofmesodiencephalic dopaminergic neurons: the role of Nurr1 and Pitx3.Adv Exp Med Biol. 2009;651:47-57. Adv Exp Med Biol. 2009. PMID: 19731549 Review. No abstract available.
Cited by
-
Lmx1b is required for the glutamatergic fates of a subset of spinal cord neurons.Neural Dev. 2016 Aug 23;11(1):16. doi: 10.1186/s13064-016-0070-1. Neural Dev. 2016. PMID: 27553035 Free PMC article.
-
ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains.Nat Methods. 2012 Jun 17;9(7):735-42. doi: 10.1038/nmeth.2076. Nat Methods. 2012. PMID: 22706672
-
Chemical Genetics Screen Identifies Epigenetic Mechanisms Involved in Dopaminergic and Noradrenergic Neurogenesis in Zebrafish.Front Genet. 2020 Feb 25;11:80. doi: 10.3389/fgene.2020.00080. eCollection 2020. Front Genet. 2020. PMID: 32158467 Free PMC article.
-
A mutation in cnot8, component of the Ccr4-not complex regulating transcript stability, affects expression levels of developmental regulators and reveals a role of Fgf3 in development of caudal hypothalamic dopaminergic neurons.PLoS One. 2014 Dec 5;9(12):e113829. doi: 10.1371/journal.pone.0113829. eCollection 2014. PLoS One. 2014. PMID: 25478689 Free PMC article.
-
Sim1a and Arnt2 contribute to hypothalamo-spinal axon guidance by regulating Robo2 activity via a Robo3-dependent mechanism.Development. 2013 Jan 1;140(1):93-106. doi: 10.1242/dev.087825. Development. 2013. PMID: 23222439 Free PMC article.
References
-
- Björklund A, Lindvall O. Dopamine containing systems in the CNS. In: Björklund A, Höckfelt T, editor. Handbook of chemical neuroanatomy: Classical Transmitters in the CNS. Vol. 2. Amsterdam: Elsevier; 1984. pp. 55–121.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

